Search Count: 22
All
Selected
 |
Crystal Structure Of Lactobacillus Rhamnosus L-Rhamnose Isomerase In Complex With L-Rhamnose
Organism: Lacticaseibacillus rhamnosus
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2024-03-13 Classification: ISOMERASE Ligands: MN, RM4, RAM |
 |
Organism: Pseudomonas syringae, Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2024-02-28 Classification: TRANSFERASE Ligands: UDP, RAM |
 |
X-Ray Structure Of A L-Rhamnose-Alpha-1,4-D-Glucuronate Lyase From Fusarium Oxysporum 12S, L-Rha Complex At 100K
Organism: Fusarium oxysporum
Method: X-RAY DIFFRACTION Resolution:1.06 Å Release Date: 2024-01-24 Classification: LYASE Ligands: RAM, ACT, TRS, CA, NA |
 |
Neutron Structure Of A L-Rhamnose-Alpha-1,4-D-Glucuronate Lyase From Fusarium Oxysporum 12S, L-Rha Complex
Organism: Fusarium oxysporum
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.25 Å, 1.8 Å Release Date: 2023-08-09 Classification: LYASE Ligands: RAM, ACT, TRS, CA, NA |
 |
Crystal Structure Of A L-Rhamnose-Alpha-1,4-D-Glucuronate Lyase From Fusarium Oxysporum 12S, L-Rha Complex
Organism: Fusarium oxysporum
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2021-08-04 Classification: LYASE Ligands: RAM, ACT, NA |
 |
Organism: Streptomyces antibioticus
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2020-10-21 Classification: OXIDOREDUCTASE Ligands: HEM, DEB, RAM, TRS, FMT, NA, GOL |
 |
Organism: Streptomyces antibioticus
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2020-10-21 Classification: OXIDOREDUCTASE Ligands: HEM, QR8, RAM, FMT, NA |
 |
Crystal Structure Of Lectin From Pleurotus Ostreatus In Complex With Rhamnose
Organism: Pleurotus ostreatus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2020-08-05 Classification: SUGAR BINDING PROTEIN Ligands: CA, RAM |
 |
Crystal Structure Of A Rhamnose-Binding Lectin Sul-I From The Toxopneustid Sea Urchin Toxopneustes Pileolus
Organism: Toxopneustes pileolus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-05-17 Classification: SUGAR BINDING PROTEIN Ligands: RAM, PO4 |
 |
Organism: Klebsiella oxytoca
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2015-04-15 Classification: HYDROLASE Ligands: RAM, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2015-03-04 Classification: IMMUNE SYSTEM Ligands: ACT, RAM |
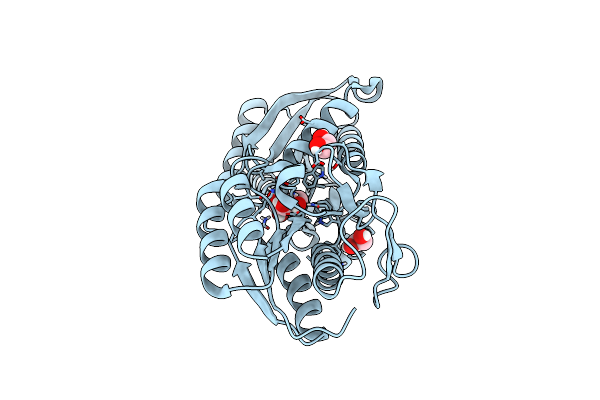 |
Crystal Structure Of An Abc Transporter Solute Binding Protein (Ipr025997) From Clostridium Phytofermentas (Cphy_0583, Target Efi-511148) With Bound L-Rhamnose
Organism: Clostridium phytofermentans
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2014-12-17 Classification: TRANSPORT PROTEIN Ligands: RAM, EDO |
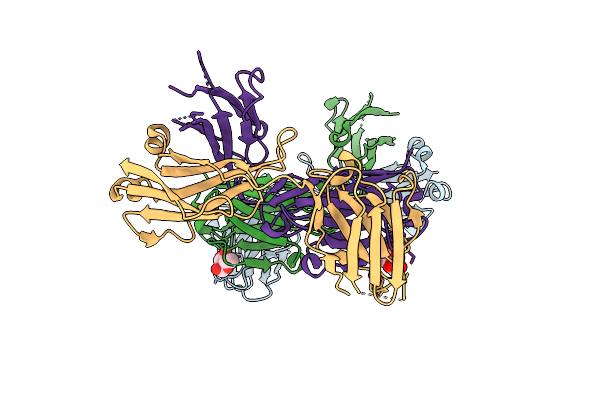 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-08-28 Classification: IMMUNE SYSTEM Ligands: RAM |
 |
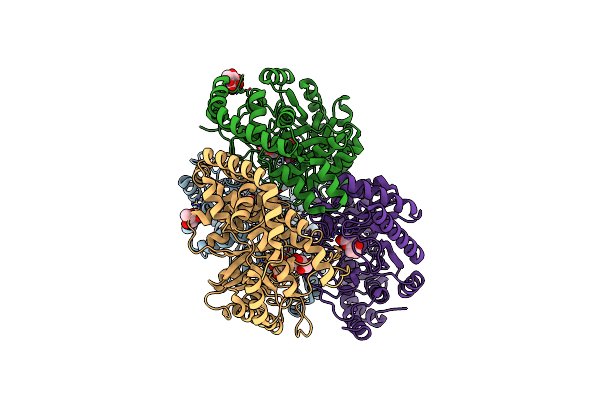
Crystal Structure Of Streptomyces Avermitilis Alpha-L-Rhamnosidase Complexed With L-Rhamnose
Organism: Streptomyces avermitilis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2013-03-20 Classification: HYDROLASE Ligands: CA, RAM, MPD, NA |
 |
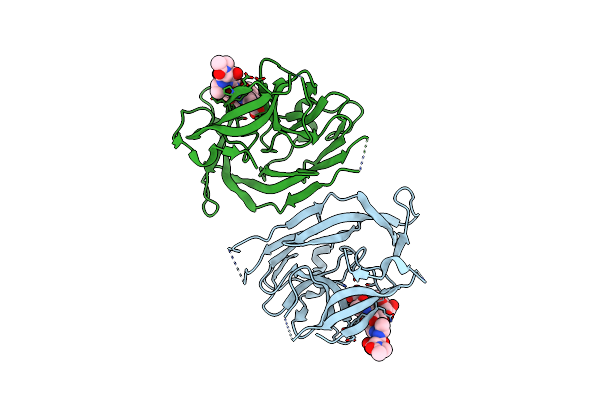
Crystal Structure Of Pseudomonas Stutzeri L-Rhamnose Isomerase Mutant H101N In Complex With L-Rhamnopyranose
Organism: Pseudomonas stutzeri
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2012-12-12 Classification: ISOMERASE Ligands: RNS, RAM, MN, RM4 |
 |
Crystal Structure Of Escherichia Coli Type I Signal Peptidase In Complex With An Arylomycin Lipoglycopeptide Antibiotic
Organism: Escherichia coli, Streptomyces sp.
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2011-10-05 Classification: HYDROLASE/ANTIBIOTIC Ligands: 02U, RAM |
 |
Crystal Structure Of The Complex Of Peptidoglycan Recognition Protein (Pgrp-S) With Alpha-Rhamnose At 2.9 A Resolution
Organism: Camelus dromedarius
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2010-07-21 Classification: ANTIBIOTIC Ligands: TLA, RAM |
 |
Organism: Oncorhynchus keta
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2009-06-30 Classification: IMMUNE SYSTEM, SUGAR BINDING PROTEIN Ligands: PO4, RAM |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.32 Å Release Date: 2009-02-03 Classification: LYASE Ligands: CA, RAM, MPD |
 |
Organism: Mus musculus
Method: SOLUTION NMR Release Date: 2008-04-29 Classification: CELL ADHESION, SIGNALING PROTEIN Ligands: NAG, RAM |

