Search Count: 56
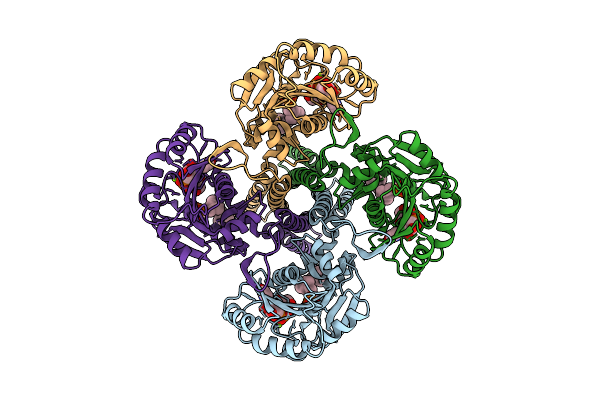 |
Cryo-Em Structure Of The Glycosyltransferase Gtrb In The Substrate-Bound State
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: UPG, 5TR, MG |
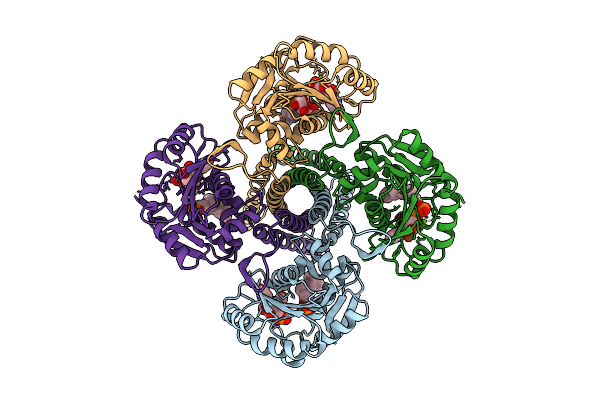 |
Cryo-Em Structure Of The Glycosyltransferase Gtrb In The Pre-Catalysis And Product-Bound State
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: UPG, 5TR, MG, UDP, A1B94 |
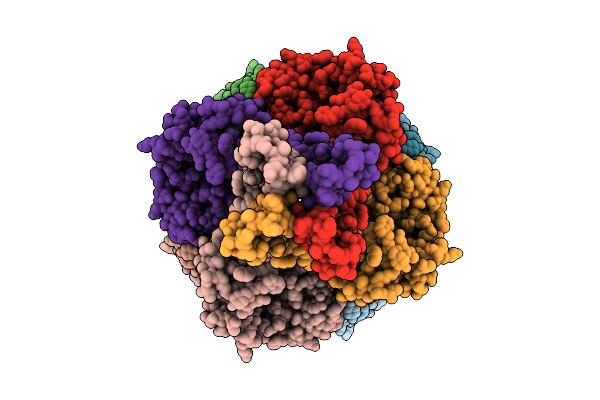 |
Cryo-Em Structure Of The Glycosyltransferase Gtrb In The Apo State (Octamer Volume)
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN |
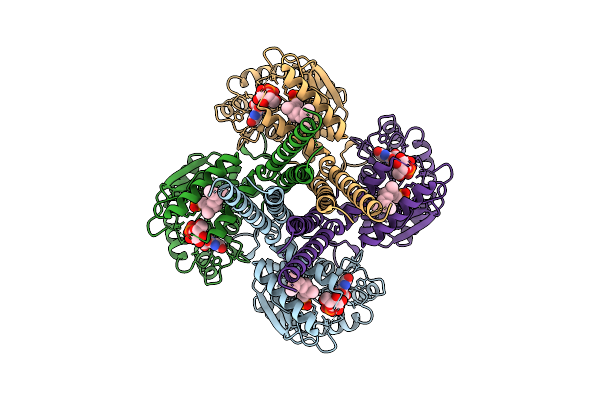 |
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: UPG, 5TR, MG |
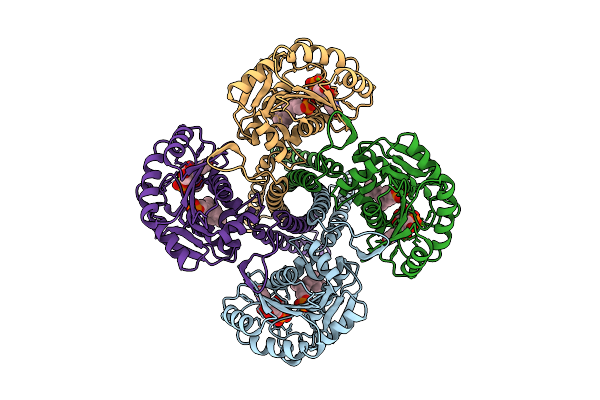 |
Cryo-Em Structure Of The Glycosyltransferase Gtrb In The Pre-Intermediate State
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: UPG, 5TR, MG |
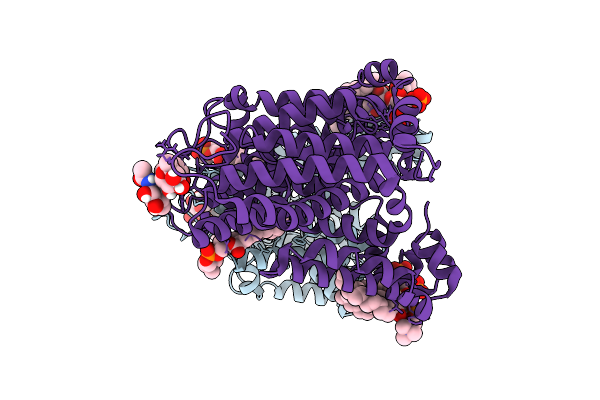 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN Ligands: PLC, PIO |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: TRANSPORT PROTEIN Ligands: PIO, PLC, NAG |
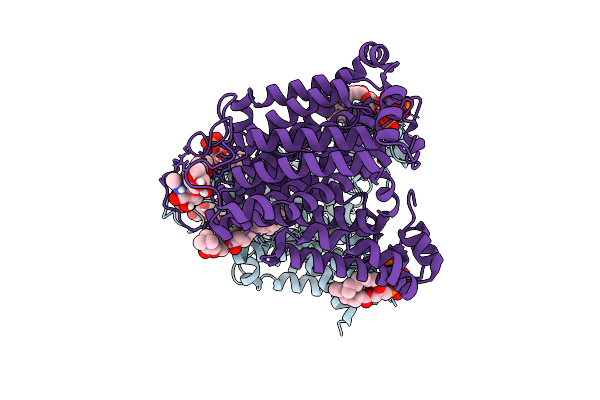 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: TRANSPORT PROTEIN Ligands: PIO, PLC, CL |
 |
Organism: Mus musculus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: TRANSPORT PROTEIN Ligands: CHT |
 |
Organism: Mus musculus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: TRANSPORT PROTEIN Ligands: CHT |
 |
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2024-02-07 Classification: TRANSPORT PROTEIN Ligands: CLR, LBN, CL, DMU, AV0 |
 |
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2024-02-07 Classification: TRANSPORT PROTEIN Ligands: AV0, IOD, CLR, LBN |
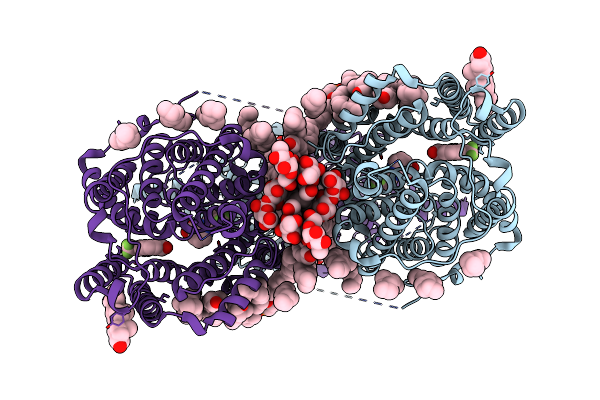 |
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2024-02-07 Classification: TRANSPORT PROTEIN Ligands: CL, LBN, NFL, AV0, CLR |
 |
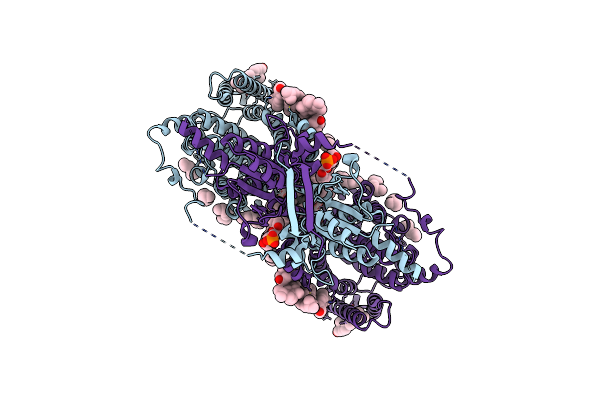
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2024-02-07 Classification: TRANSPORT PROTEIN Ligands: CLR, LBN, BCT, DMU, AV0 |
 |
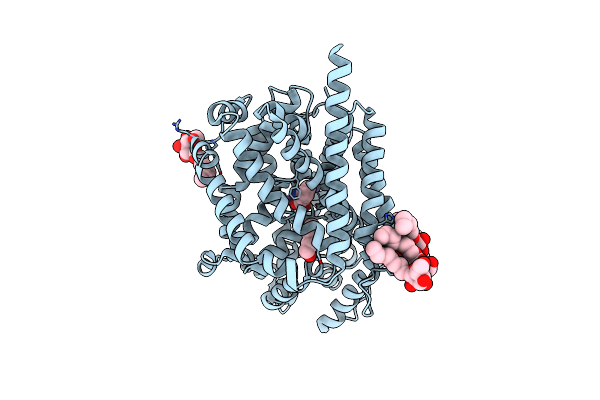
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2024-02-07 Classification: TRANSPORT PROTEIN Ligands: CLR, LBN, AV0 |
 |
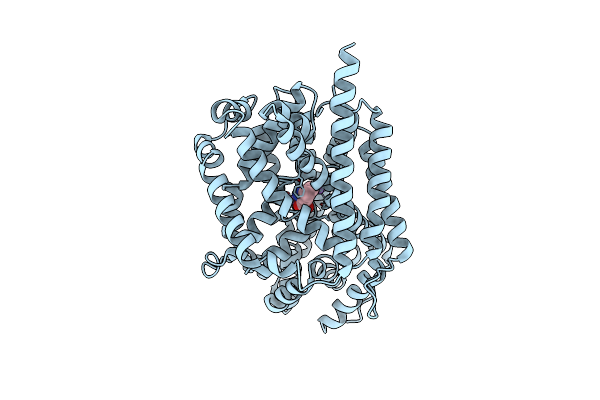
Organism: Aquifex aeolicus
Method: X-RAY DIFFRACTION Resolution:4.00 Å Release Date: 2023-11-22 Classification: TRANSPORT PROTEIN |
 |
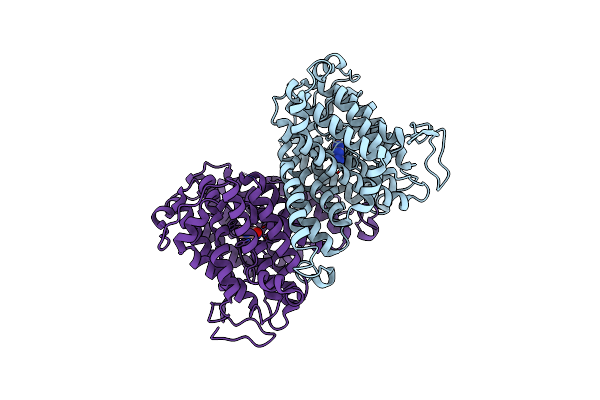
Organism: Aquifex aeolicus
Method: X-RAY DIFFRACTION Resolution:3.80 Å Release Date: 2023-11-22 Classification: TRANSPORT PROTEIN |
 |
Organism: Colwellia psychrerythraea
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2023-01-25 Classification: TRANSPORT PROTEIN Ligands: GUN |
 |
Organism: Aquifex aeolicus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2021-04-07 Classification: TRANSPORT PROTEIN Ligands: LEU, NA |
 |
Organism: Aquifex aeolicus
Method: X-RAY DIFFRACTION Resolution:3.49 Å Release Date: 2021-04-07 Classification: TRANSPORT PROTEIN Ligands: MSE, NA |

