Search Count: 4
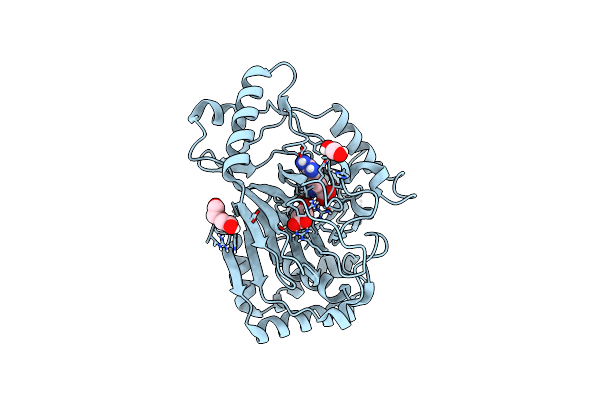 |
Crystal Structure Of The L-Arginine Hydroxylase Vioc Mehis316, Bound To Fe(Ii), L-Arginine, And Succinate
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-07-31 Classification: OXIDOREDUCTASE Ligands: FE2, SIN, ARG, EDO, PEG |
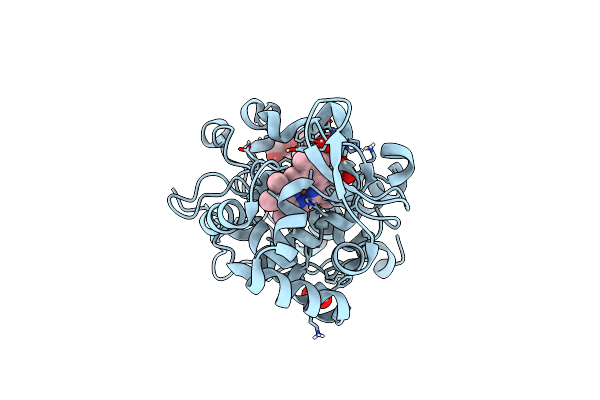 |
The Crystal Structure Of Engineered Cytochrome C Peroxidase From Saccharomyces Cerevisiae With A Trp51 To S-Trp51 Modification
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2021-06-16 Classification: OXIDOREDUCTASE Ligands: HEM, EDO, 1PE |
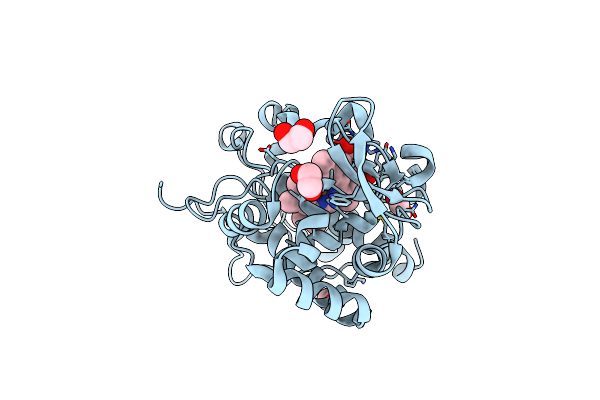 |
The Crystal Structure Of Engineered Cytochrome C Peroxidase From Saccharomyces Cerevisiae With Trp51 To S-Trp51 And Trp191Phe Modifications
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-06-16 Classification: OXIDOREDUCTASE Ligands: HEM, EDO |
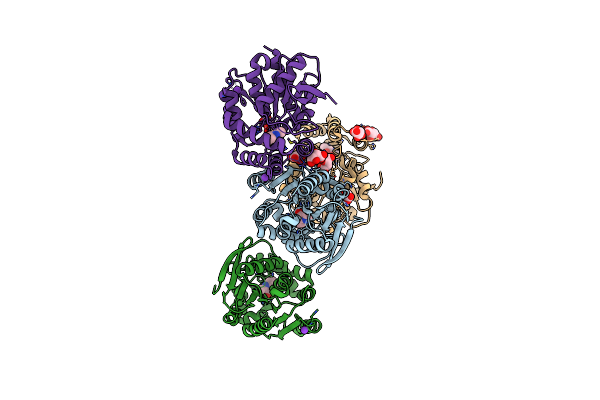 |
Crystal Structure Of The Cofactor-Devoid 1-H-3-Hydroxy-4- Oxoquinaldine 2,4-Dioxygenase (Hod) Catalytically Inactive H251A Variant Complexed With Its Natural Substrate 1-H-3-Hydroxy-4- Oxoquinaldine
Organism: Arthrobacter nitroguajacolicus
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2013-12-04 Classification: OXIDOREDUCTASE Ligands: K, HQD, TAR |

