Search Count: 66
 |
Cryo-Em Structure Of Enterovirus A71 Mature Virion In Complex With Fab H1A6.2
Organism: Enterovirus a71
Method: ELECTRON MICROSCOPY Release Date: 2024-09-04 Classification: VIRUS Ligands: SPH |
 |
Organism: Enterovirus a71
Method: ELECTRON MICROSCOPY Release Date: 2024-09-04 Classification: VIRUS |
 |
Cryo-Em Structure Of Enterovirus A71 Empty Particle In Complex With Fab H1A6.2
Organism: Enterovirus a71
Method: ELECTRON MICROSCOPY Release Date: 2024-09-04 Classification: VIRUS |
 |
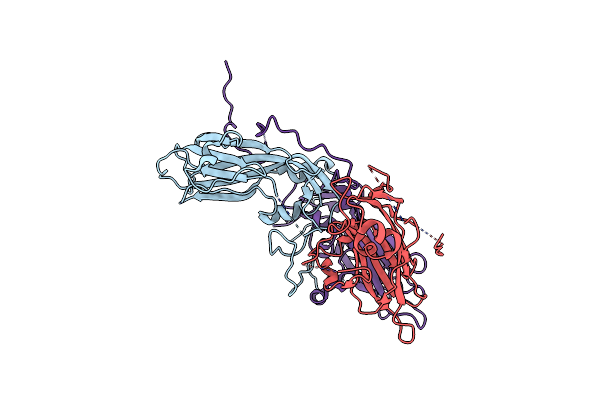
Cryo-Em Structure Of Coxsackievirus A16 Mature Virion In Complex With Fab H1A6.2
Organism: Coxsackievirus a16
Method: ELECTRON MICROSCOPY Release Date: 2024-09-04 Classification: VIRUS |
 |
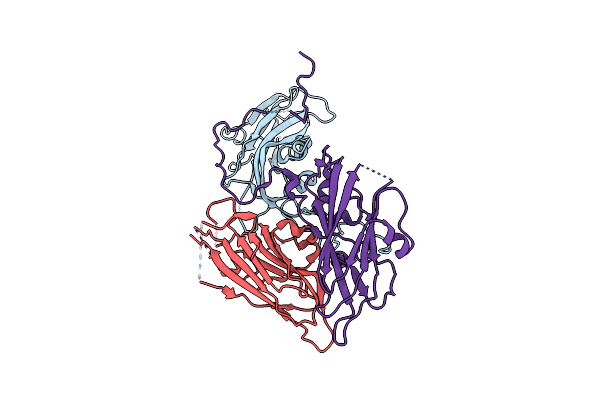
Cryo-Em Structure Of Coxsackievirus A16 A-Particle In Complex With Fab H1A6.2
Organism: Coxsackievirus a16
Method: ELECTRON MICROSCOPY Release Date: 2024-09-04 Classification: VIRUS |
 |
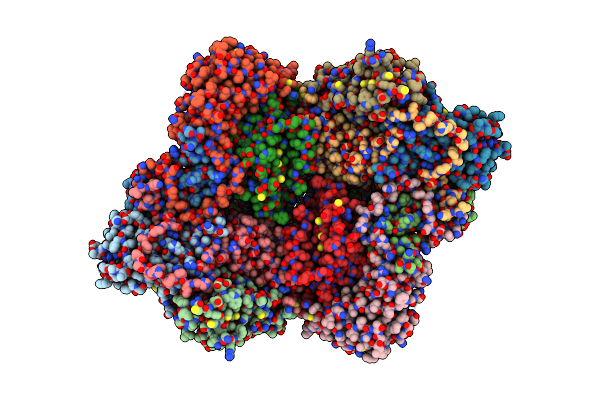
Cryo-Em Structure Of Coxsackievirus A16 Empty Particle In Complex With Fab H1A6.2
Organism: Coxsackievirus a16
Method: ELECTRON MICROSCOPY Release Date: 2024-09-04 Classification: VIRUS |
 |
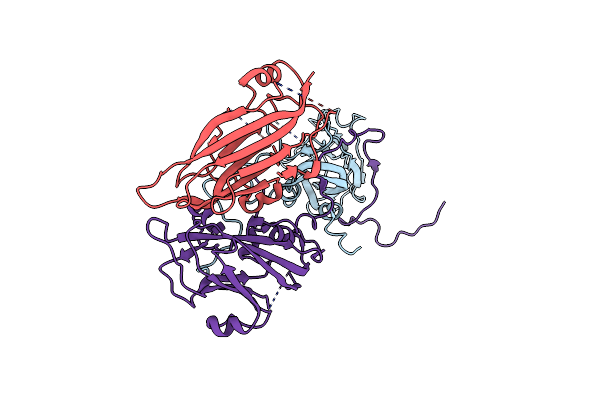
Cryo-Em Structure Of Coxsackievirus A16 Empty Particle In Complex With Fab H1A6.2 (Local Refinement)
Organism: Mus musculus, Coxsackievirus a16
Method: ELECTRON MICROSCOPY Release Date: 2024-09-04 Classification: VIRAL PROTEIN |
 |
Organism: Enterovirus a71
Method: ELECTRON MICROSCOPY Release Date: 2024-09-04 Classification: VIRUS |
 |
Organism: Enterovirus a71
Method: ELECTRON MICROSCOPY Release Date: 2024-09-04 Classification: VIRUS Ligands: SPH |
 |
Cryo-Em Structure Of Coxsackievirus B1 A-Particle In Complex With Nab 8A10 (Cvb1-A:8A10)
Organism: Coxsackievirus b1, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2024-07-24 Classification: VIRUS |
 |
Cryo-Em Structure Of Sars-Cov-2 Spike Protein In Complex With Double Nabs 8H12 And 3E2 (Local Refinement)
Organism: Severe acute respiratory syndrome coronavirus 2, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2023-08-16 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Cryo-Em Structure Of Sars-Cov-2 Spike Protein In Complex With Double Nabs 8H12 And 1C4 (Local Refinement)
Organism: Severe acute respiratory syndrome coronavirus 2, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2023-08-16 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Cryo-Em Structure Of Sars-Cov-2 Spike Protein In Complex With Double Nabs 3E2 And 1C4 (Local Refinement)
Organism: Severe acute respiratory syndrome coronavirus 2, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2023-08-16 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Cryo-Em Structure Of Sars-Cov-2 Spike Protein In Complex With Double Nabs Xma01 And 3E2 (Local Refinement)
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2023-08-16 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Organism: Dinoroseobacter phage vb_dshs-r4c
Method: ELECTRON MICROSCOPY Release Date: 2023-07-12 Classification: VIRAL PROTEIN |
 |
Organism: Dinoroseobacter phage vb_dshs-r4c
Method: ELECTRON MICROSCOPY Release Date: 2023-07-12 Classification: VIRAL PROTEIN |
 |
Organism: Dinoroseobacter phage vb_dshs-r4c
Method: ELECTRON MICROSCOPY Release Date: 2023-07-12 Classification: VIRAL PROTEIN |
 |
Organism: Dinoroseobacter phage vb_dshs-r4c
Method: ELECTRON MICROSCOPY Release Date: 2023-07-12 Classification: VIRUS |
 |
Cryo-Em Model Of The Marine Siphophage Vb_Dshs-R4C Stopper-Terminator Complex
Organism: Dinoroseobacter phage vb_dshs-r4c
Method: ELECTRON MICROSCOPY Release Date: 2023-07-12 Classification: VIRUS |
 |
Cryo-Em Structure Of Coxsackievirus B1 Empty Particle In Complex With Nab 9A3 (Cvb1-E:9A3)
Organism: Mus musculus, Coxsackievirus b1
Method: ELECTRON MICROSCOPY Release Date: 2023-06-07 Classification: VIRUS |

