Search Count: 24
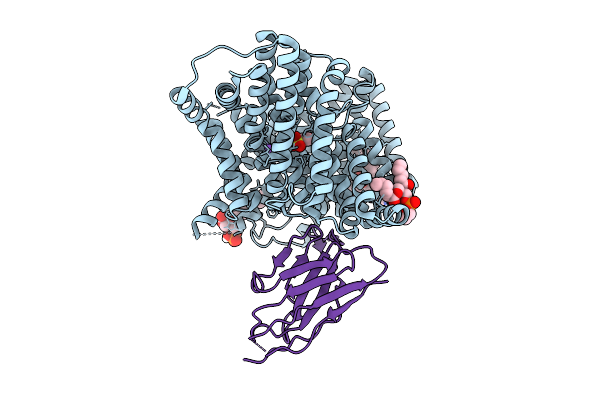 |
Cryo-Em Structure Of The Isethionate Trap Transporter Iseqm From Oleidesulfovibrio Alaskensis With Bound Isethionate
Organism: Oleidesulfovibrio alaskensis g20, Helicobacter pylori
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: TRANSPORT PROTEIN |
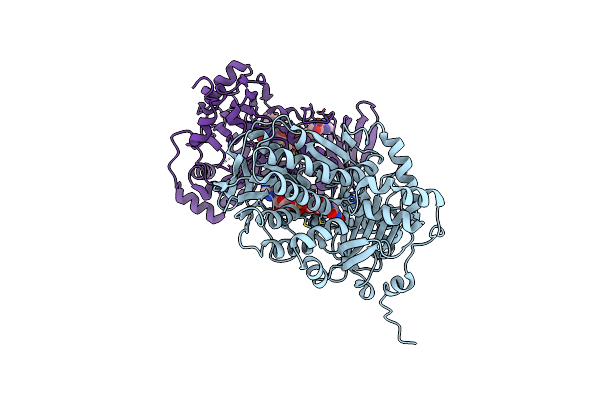 |
Organism: Streptomyces lasalocidi
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-12-13 Classification: FLAVOPROTEIN Ligands: FAD, CL |
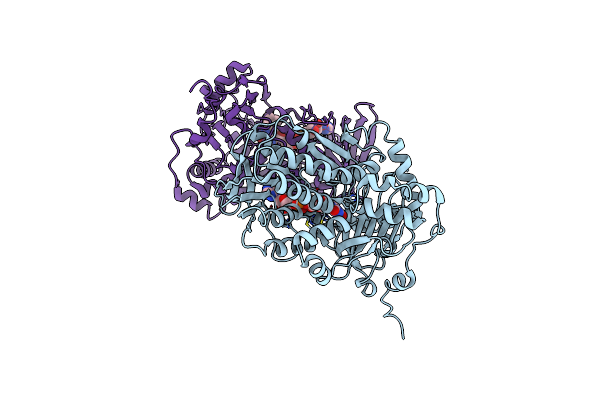 |
Organism: Streptomyces lasalocidi
Method: X-RAY DIFFRACTION Resolution:3.76 Å Release Date: 2023-12-13 Classification: FLAVOPROTEIN Ligands: FAD |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2022-10-05 Classification: HYDROLASE Ligands: D06 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2022-10-05 Classification: HYDROLASE Ligands: D06, TMO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.78 Å Release Date: 2022-10-05 Classification: HYDROLASE Ligands: CW8 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2022-10-05 Classification: HYDROLASE Ligands: CW8, TMO |
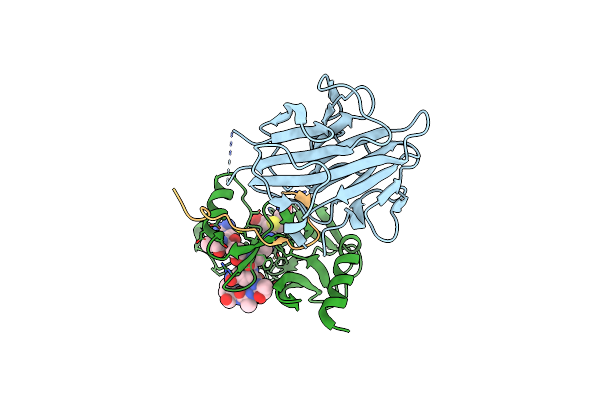 |
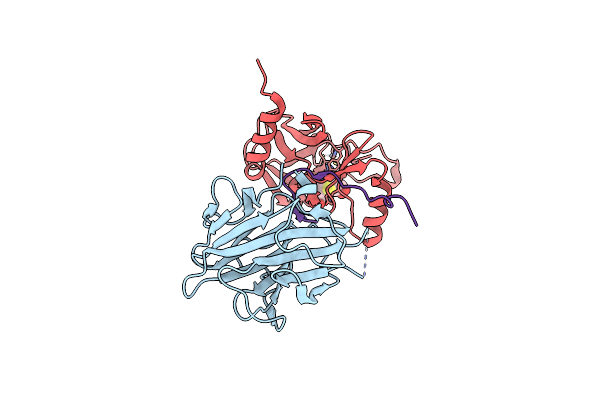
The Crystal Structure Of Mll1 (N3861I/Q3867L/C3882Ss)-Rbbp5-Ash2L In Complex With H3K4Me0 Peptide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2022-09-07 Classification: PROTEIN BINDING Ligands: SAH, ZN |
 |
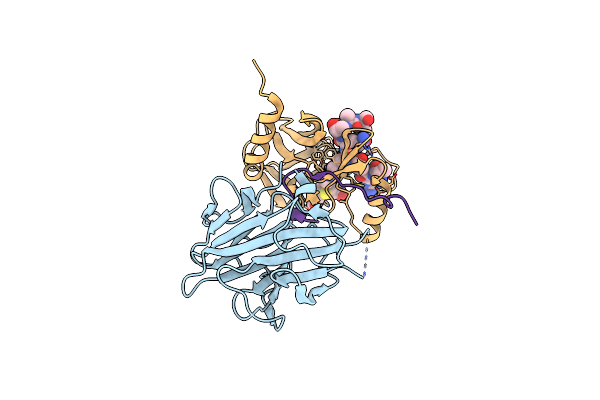
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2022-09-07 Classification: PROTEIN BINDING Ligands: SAH, ZN |
 |
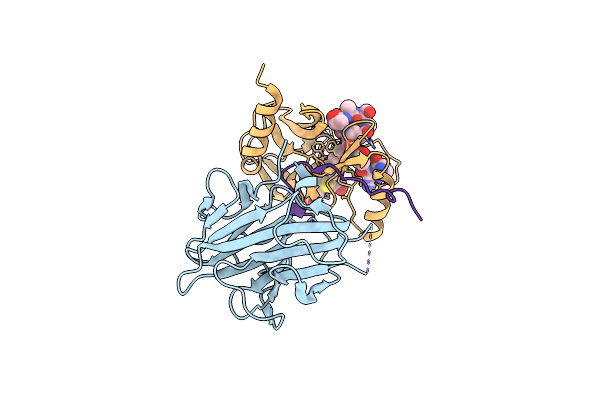
The Crystal Structure Of Mll1 (N3861I/Q3867L/C3882Ss)-Rbbp5-Ash2L In Complex With H3K4Me1 Peptide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.56 Å Release Date: 2022-09-07 Classification: PROTEIN BINDING Ligands: SAH, ZN |
 |
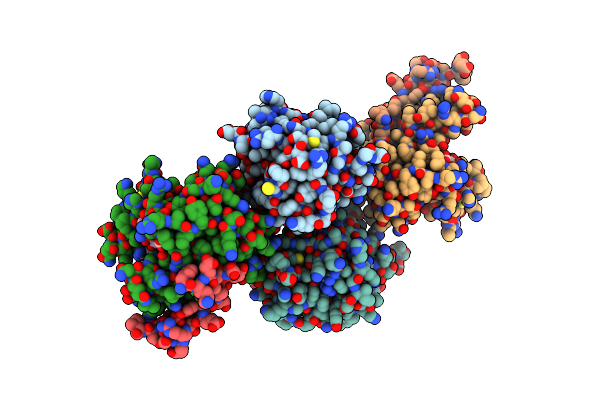
The Crystal Structure Of Mll1 (N3861I/Q3867L/C3882Ss)-Rbbp5-Ash2L In Complex With H3K4Me2 Peptide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2022-09-07 Classification: PROTEIN BINDING Ligands: SAH, ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2022-09-07 Classification: PROTEIN BINDING/TRANSFERASE Ligands: ZN, SAH |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: SOLUTION NMR Release Date: 2020-08-05 Classification: TRANSCRIPTION |
 |
Crystal Structure Of Atcpsf30 Yth Domain In Complex With 10Mer M6A-Modified Rna
Organism: Arabidopsis thaliana, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2019-05-15 Classification: RNA BINDING PROTEIN/RNA Ligands: GOL, EPE |
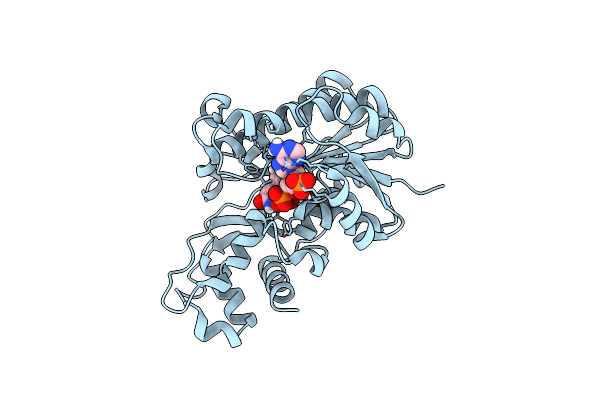 |
Structure-Guided Engineering Of Reductase: Efficient Attenuating Substrate Inhibition In Asymmetric Catalysis
Organism: Scheffersomyces stipitis
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2019-03-06 Classification: OXIDOREDUCTASE Ligands: NAP |
 |
Organism: Scheffersomyces stipitis cbs 6054
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-03-06 Classification: OXIDOREDUCTASE |
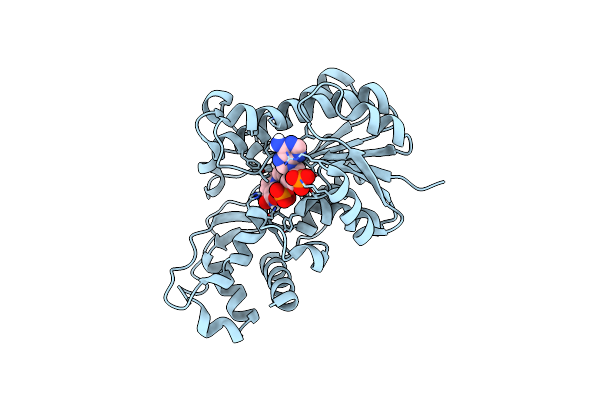 |
Organism: Scheffersomyces stipitis (strain atcc 58785 / cbs 6054 / nbrc 10063 / nrrl y-11545)
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2019-03-06 Classification: OXIDOREDUCTASE Ligands: NAP |
 |
Organism: Edwardsiella tarda (strain eib202)
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2019-03-06 Classification: OXIDOREDUCTASE Ligands: SO4 |
 |
Nmr Structural And Biophysical Functional Analysis Of Intracellular Loop 5 Of The Nhe1 Isoform Of The Na+/H+ Exchanger.
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2018-11-14 Classification: MEMBRANE PROTEIN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2017-08-16 Classification: CHAPERONE Ligands: IMD, ZN, CL |

