Search Count: 8
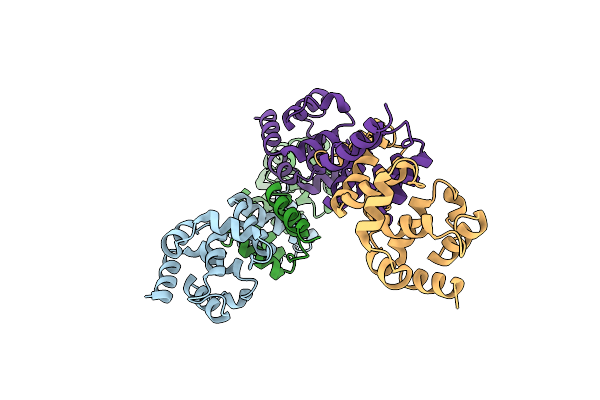 |
Structural Basis For Dimerization Of The Death Effector Domains Of Caspase-8
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.17 Å Release Date: 2017-10-25 Classification: HYDROLASE |
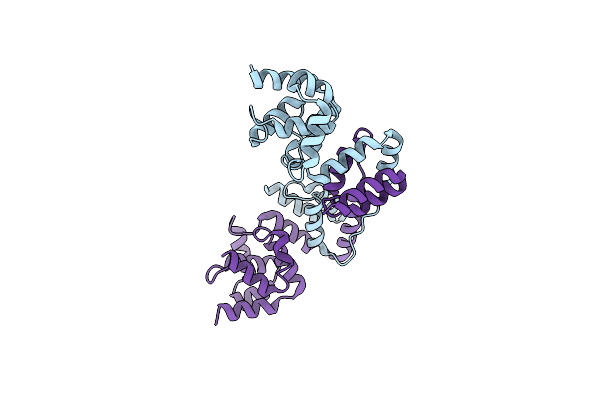 |
Structural Basis For Dimerization Of The Death Effector Domains Of Caspase-8
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2017-10-25 Classification: HYDROLASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2015-07-01 Classification: HYDROLASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-12-24 Classification: TRANSFERASE/PEPTIDE Ligands: PO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2014-12-03 Classification: TRANSFERASE |
 |
Organism: Danio rerio, Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2013-07-24 Classification: TRANSFERASE |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-09-26 Classification: HYDROLASE Ligands: ACT, ZN |
 |
Crystal Structure Of Imidazolonepropionase Complexed With Imidazole-4-Acetic Acid Sodium Salt, A Substrate Homologue
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-09-26 Classification: HYDROLASE Ligands: ZN, IZC |

