Search Count: 80
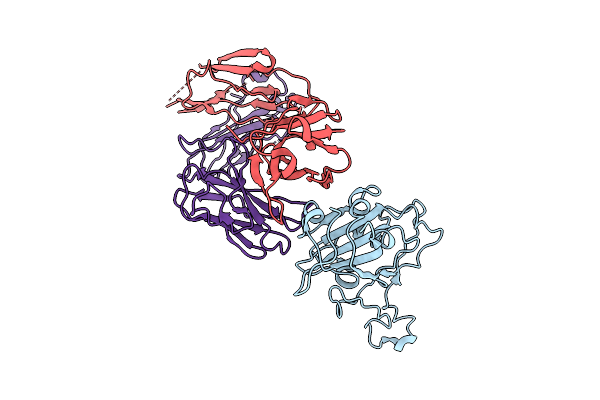 |
The Local Refined Map Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-171
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
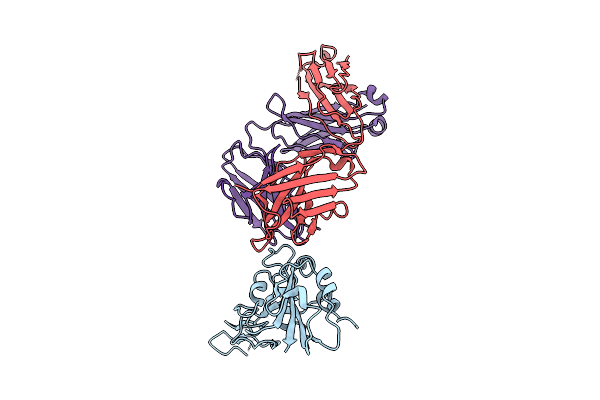
The Local Refined Map Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-183
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
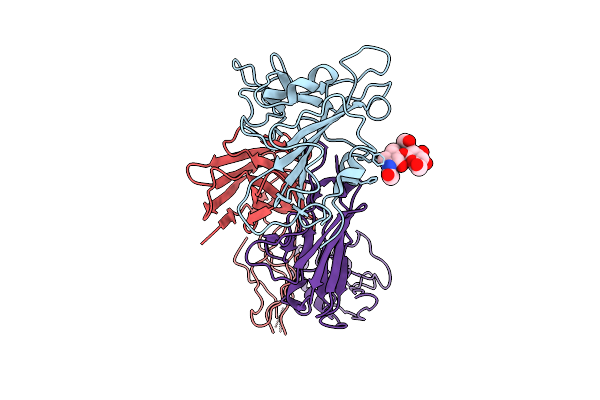
Structure Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-171
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
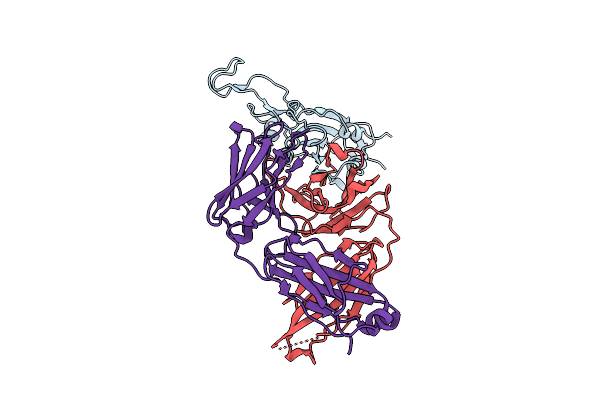
The Local Refined Map Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-198
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
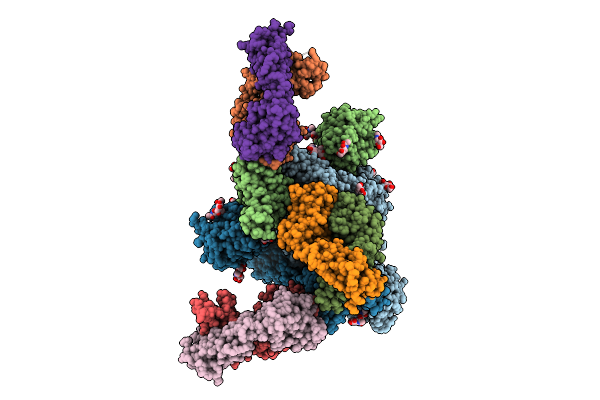
The Local Refined Map Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-203
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
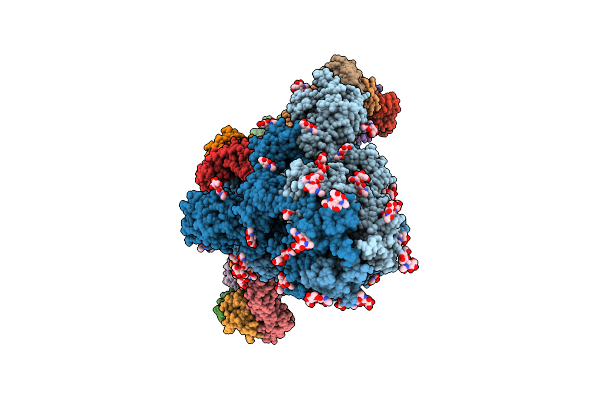
Structure Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-203
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
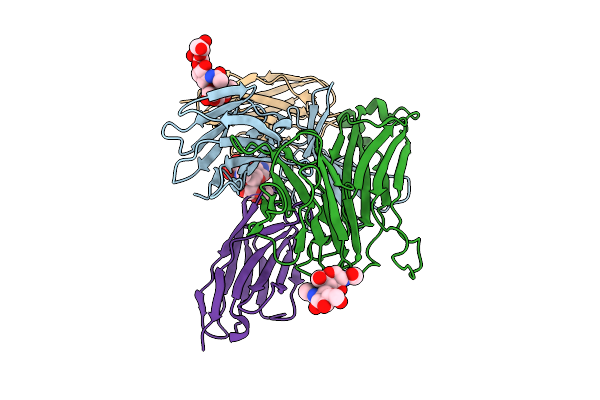
Structure Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-198
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Abtcr Bound To Sar444200, A Novel Anti-Gpc3 T-Cell Engager, For The Treatment Of Gpc3+ Solid Tumors
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: IMMUNE SYSTEM |
 |
Identification And Non-Clinical Characterization Of Sar444200, A Novel Anti-Gpc3 T-Cell Engager, For The Treatment Of Gpc3+ Solid Tumors
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: ONCOPROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
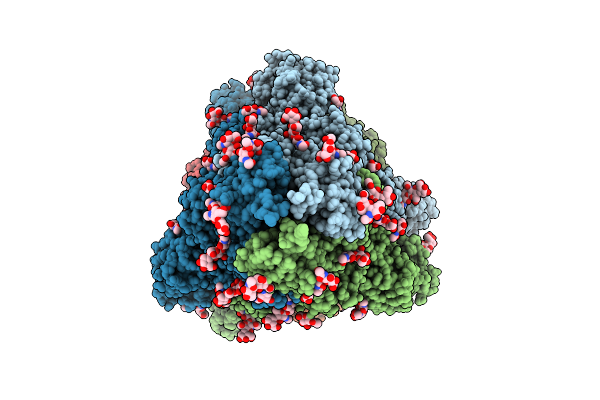
Organism: Tribonema minus
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: PHOTOSYNTHESIS Ligands: CLA, BCR, DD6, LMT, LHG, A1L1G, LMG, PQN, SF4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: LIGASE Ligands: A1EMT |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2025-05-07 Classification: LIGASE Ligands: A1D9F |
 |
Organism: Lama glama
Method: X-RAY DIFFRACTION Release Date: 2025-04-16 Classification: ANTITUMOR PROTEIN Ligands: SO4, GOL, ACT |
 |
The L-Tryptophan Specific Decarboxylase Psid(50-439) In Complex With Tryptamine
Organism: Psilocybe cubensis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-04-02 Classification: OXIDOREDUCTASE Ligands: TSS |
 |
Structure Complex Of 4-Hydroxytryptamine Kinase Psik Complexed With Mg2+ And Tryptamine
Organism: Psilocybe cubensis
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2025-04-02 Classification: METAL BINDING PROTEIN Ligands: TSS, ADP, MG, CA |
 |
Organism: Psilocybe cubensis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-04-02 Classification: TRANSFERASE |
 |
Crystal Structure Of The Methyltransferase Psim In Complex With S-Adenosylmethionine (Sam)
Organism: Psilocybe cubensis
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2025-04-02 Classification: TRANSFERASE Ligands: SAM |
 |
Organism: Psilocybe cubensis
Method: X-RAY DIFFRACTION Resolution:1.31 Å Release Date: 2025-04-02 Classification: TRANSFERASE Ligands: SAH |
 |
Crystal Structure Of The Methyltransferase Psim In Complex With Sah And Norbaeocystin
Organism: Psilocybe cubensis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2025-04-02 Classification: TRANSFERASE Ligands: SAH, XP6 |
 |
Crystal Structure Of The Methyltransferase Psim In Complex With Sah And Baeocystin
Organism: Psilocybe cubensis
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2025-04-02 Classification: TRANSFERASE Ligands: EPE, GOL, XPN, SAH |

