Search Count: 19
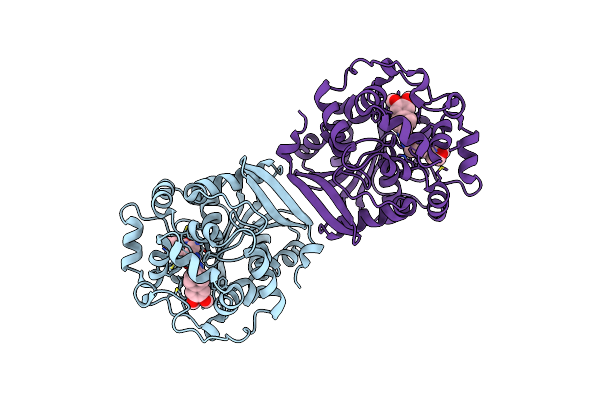 |
Crystal Structure Of Human Soluble Epoxide Hydrolase C-Terminal Domain In Complex With A Benzohomoadamantane-Based Urea Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-04-30 Classification: HYDROLASE Ligands: A1H8Y |
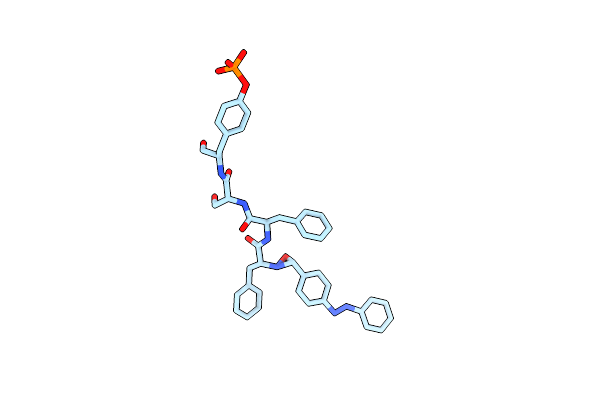 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: PROTEIN FIBRIL |
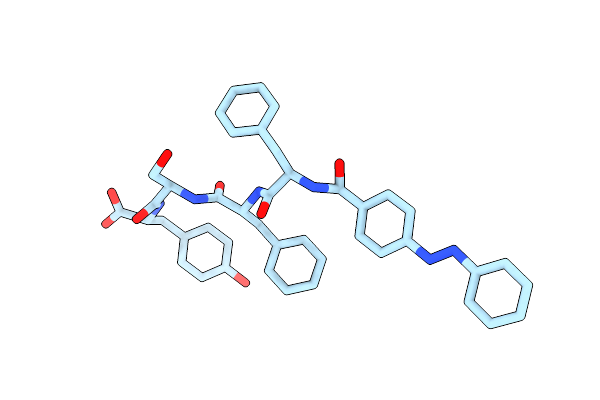 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: PROTEIN FIBRIL |
 |
Organism: Rattus norvegicus, Gallus gallus, Sus scrofa
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2021-06-30 Classification: STRUCTURAL PROTEIN Ligands: GTP, MG, CA, GOL, GDP, MES, AF6, ACP |
 |
Organism: Rattus norvegicus, Gallus gallus, Sus scrofa
Method: X-RAY DIFFRACTION Resolution:2.66 Å Release Date: 2021-06-23 Classification: STRUCTURAL PROTEIN Ligands: GTP, MG, CA, GOL, GDP, MES, AEU, ACP |
 |
Organism: Rattus norvegicus, Gallus gallus, Sus scrofa
Method: X-RAY DIFFRACTION Resolution:2.73 Å Release Date: 2021-06-23 Classification: STRUCTURAL PROTEIN Ligands: GTP, MG, CA, GOL, GDP, MES, AEX, ACP |
 |
Organism: Rattus norvegicus, Gallus gallus, Sus scrofa
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2021-06-23 Classification: STRUCTURAL PROTEIN-INHIBITOR COMPLEX Ligands: GTP, MG, CA, GDP, MES, FW9, CL, ACP |
 |
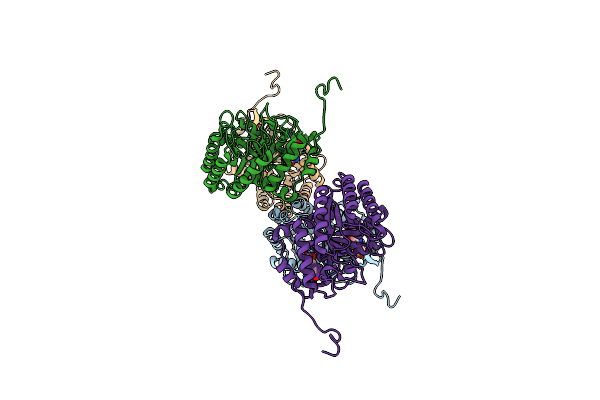
Molecular Basis For Feedback Inhibition Of Tyrosine-Regulated 3-Deoxy-D-Arabino-Heptulosonate-7-Phosphate Synthase From Escherichia Coli
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-04-17 Classification: BIOSYNTHETIC PROTEIN |
 |
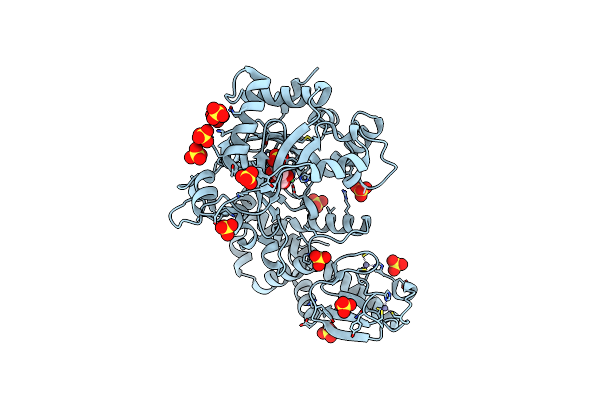
Molecular Basis For Feedback Inhibition Of Tyrosine-Regulated 3-Deoxy-D-Arabino-Heptulosonate-7-Phosphate Synthase From Escherichia Coli
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-04-17 Classification: BIOSYNTHETIC PROTEIN Ligands: TYR |
 |
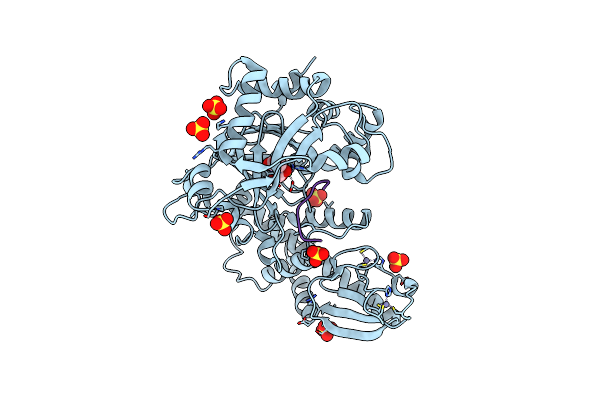
Crystal Structure Of Arabidopsis Thaliana Jmj13 Catalytic Domain In Complex With Akg
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2019-04-10 Classification: GENE REGULATION Ligands: AKG, NI, ZN, SO4 |
 |
Crystal Structure Of Arabidopsis Thaliana Jmj13 Catalytic Domain In Complex With Nog And An H3K27Me3 Peptide
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2019-04-10 Classification: GENE REGULATION Ligands: NI, ZN, OGA, SO4 |
 |
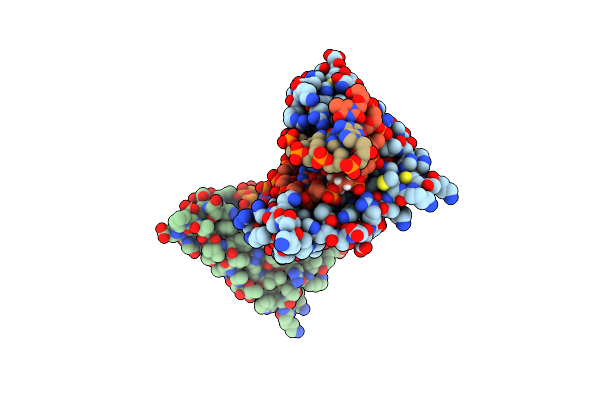
Organism: Arabidopsis thaliana, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2019-03-27 Classification: DNA BINDING PROTEIN/DNA Ligands: ZN, MG, GOL, D5M |
 |
Organism: Arabidopsis thaliana, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2019-03-27 Classification: DNA BINDING PROTEIN/DNA Ligands: ZN |
 |
Organism: Arabidopsis thaliana, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2019-03-27 Classification: DNA BINDING PROTEIN/DNA Ligands: ZN |
 |
Organism: Sus scrofa, Rattus norvegicus, Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2018-04-11 Classification: STRUCTURAL PROTEIN Ligands: GTP, MG, CA, GOL, GDP, MES, 8X0, ACP |
 |
Organism: Sus scrofa, Rattus norvegicus, Gallus gallus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2018-04-11 Classification: STRUCTURAL PROTEIN Ligands: GTP, MG, CA, GOL, GDP, MES, Y50, ACP |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-12-27 Classification: GENE REGULATION Ligands: NI, ZN |
 |
Crystal Structure Of Arabidopsis Thaliana Jmj14 Catalytic Domain In Complex With Nog And H3K4Me3 Peptide
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2017-12-27 Classification: GENE REGULATION Ligands: NI, ZN, OGA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2011-11-02 Classification: TRANSPORT PROTEIN Ligands: MYR |

