Search Count: 77
 |
Organism: Goose parvovirus
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: VIRUS LIKE PARTICLE |
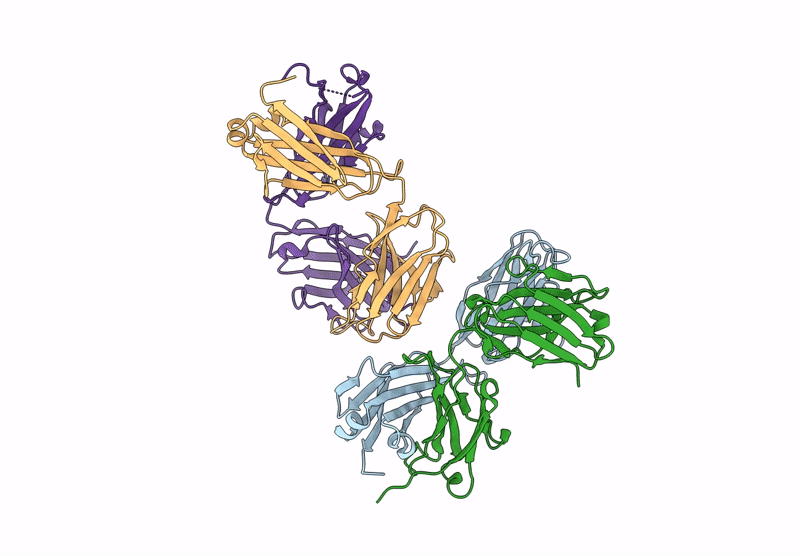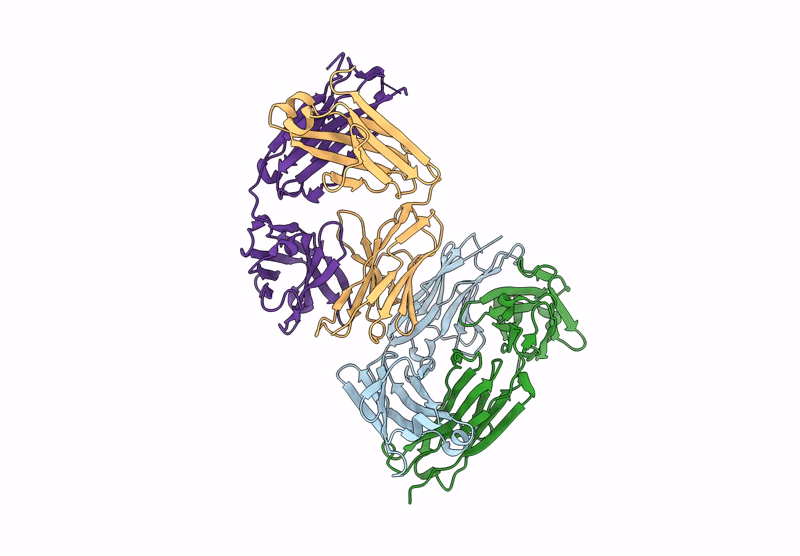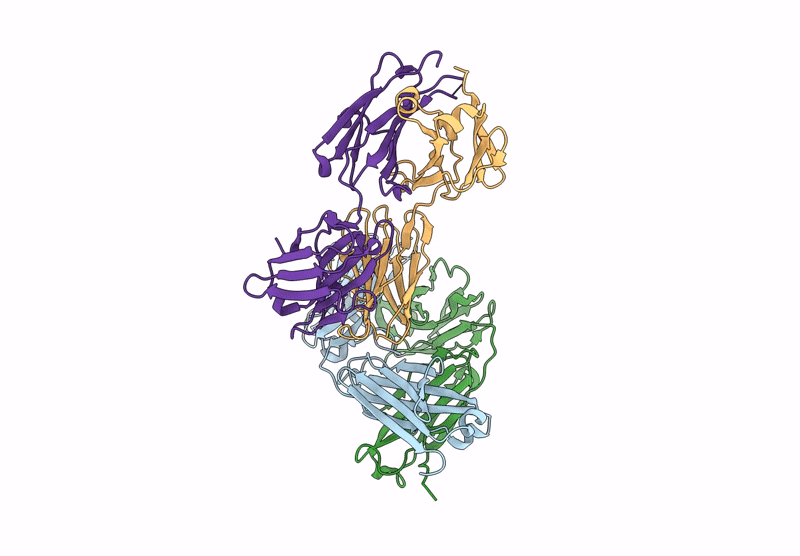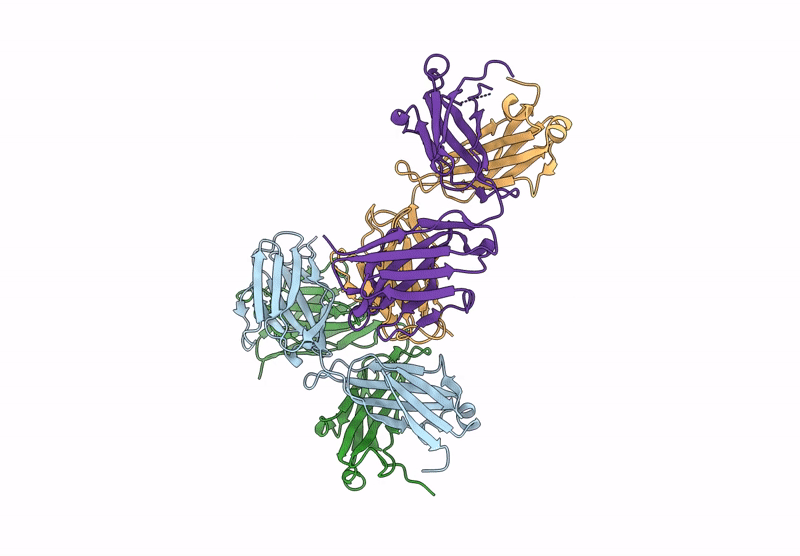 |
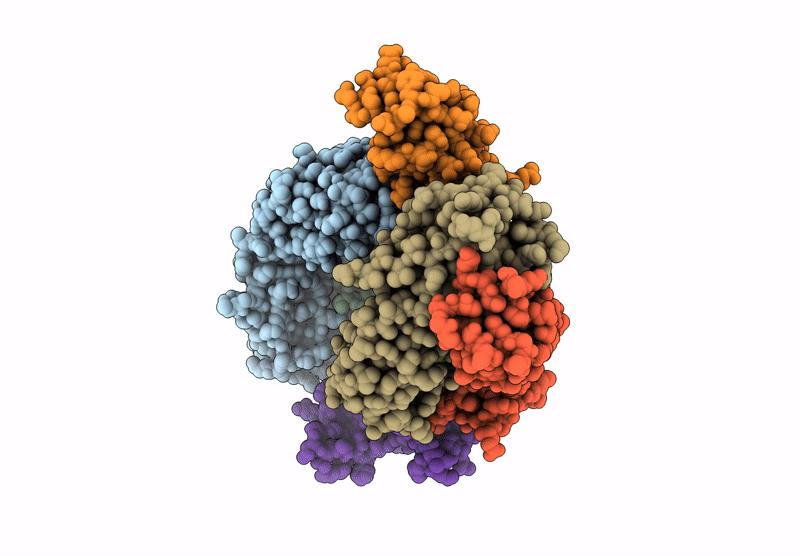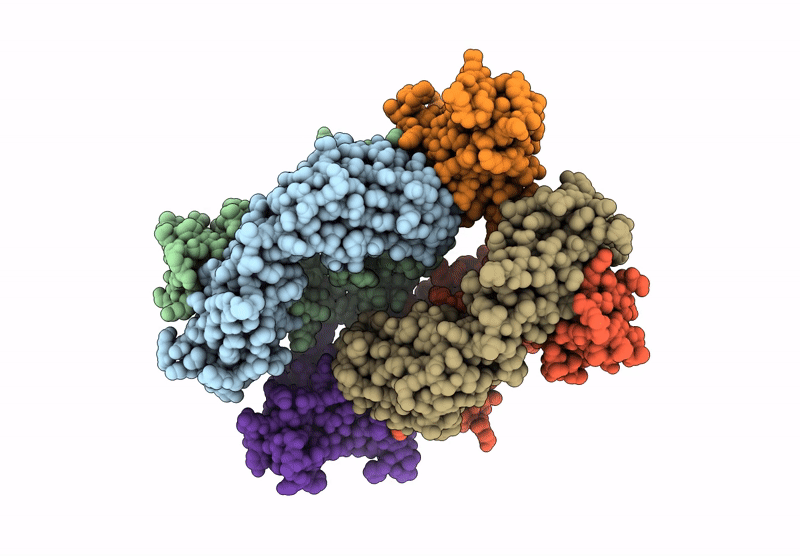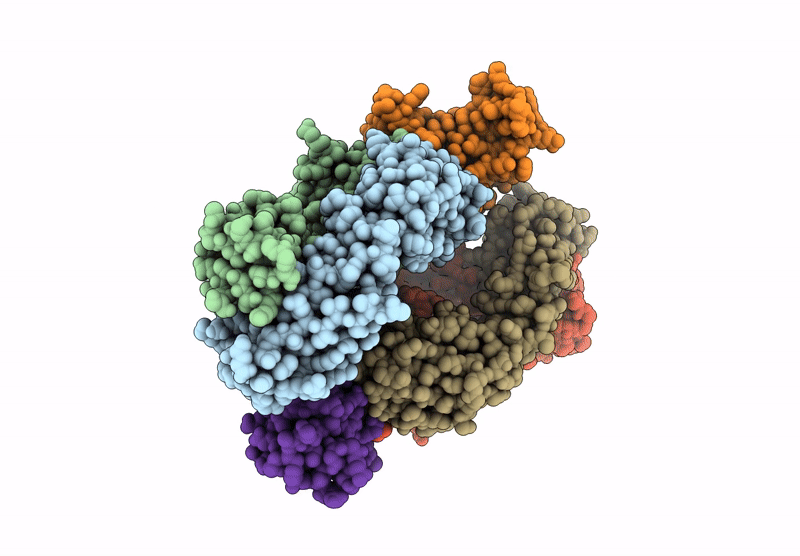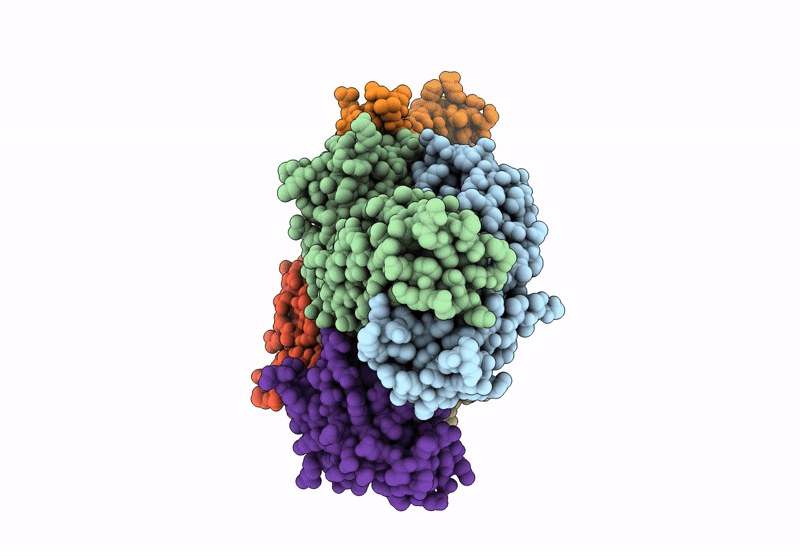
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.87 Å Release Date: 2024-12-04 Classification: IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2024-12-04 Classification: IMMUNE SYSTEM |
 |
Organism: Human papillomavirus 16, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-06-05 Classification: LIGASE/ONCOPROTEIN Ligands: ZN |
 |
Organism: Human papillomavirus 16, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-06-05 Classification: LIGASE/ONCOPROTEIN Ligands: ZN |
 |
Organism: Human papillomavirus 16, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-06-05 Classification: LIGASE/ONCOPROTEIN Ligands: ZN |
 |
Organism: Human papillomavirus 16, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-06-05 Classification: LIGASE/ONCOPROTEIN Ligands: ZN |
 |
Organism: Homo sapiens, Human papillomavirus 16
Method: ELECTRON MICROSCOPY Release Date: 2024-06-05 Classification: LIGASE/ONCOPROTEIN Ligands: ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2024-05-29 Classification: OXIDOREDUCTASE Ligands: SO4, PG4, FMN, OG6, ACT, CL, FJW |
 |
Structure Of An Orthogonal Pyr1*:Hab1* Chemical-Induced Dimerization Module In Complex With Mandipropamid
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2023-11-08 Classification: PLANT PROTEIN Ligands: GOL, 3UZ |
 |
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2023-10-25 Classification: ANTIBIOTIC |
 |
Organism: Hansschlegelia zhihuaiae
Method: X-RAY DIFFRACTION Resolution:1.29 Å Release Date: 2023-08-02 Classification: HYDROLASE Ligands: 1MM, GOL |
 |
Organism: Hansschlegelia zhihuaiae
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-08-02 Classification: HYDROLASE Ligands: IJC, GOL |
 |
Organism: Hansschlegelia zhihuaiae
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2023-08-02 Classification: HYDROLASE Ligands: GOL, JV6 |
 |
Organism: Hansschlegelia zhihuaiae
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2023-08-02 Classification: HYDROLASE Ligands: GOL, CIT |
 |
Organism: Hansschlegelia zhihuaiae
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2023-08-02 Classification: HYDROLASE Ligands: CIE, GOL |
 |
Organism: Hansschlegelia zhihuaiae
Method: X-RAY DIFFRACTION Resolution:1.32 Å Release Date: 2023-08-02 Classification: HYDROLASE Ligands: 1MM, GOL, TLA |
 |
Organism: Hansschlegelia zhihuaiae
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-08-02 Classification: HYDROLASE Ligands: 1MM, TLA, GOL |
 |
Organism: Hansschlegelia zhihuaiae
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2023-08-02 Classification: HYDROLASE Ligands: RXF, TLA, GOL |
 |
Organism: Hansschlegelia zhihuaiae
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2023-08-02 Classification: HYDROLASE Ligands: 1TB, TLA, GOL |

