Search Count: 50
 |
Organism: Pyrinomonas methylaliphatogenes
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Sphingomonas sp. tpd3009
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: HYDROLASE |
 |
Dimeric Structure Of Full-Length Crga, A Cell Division Protein From Mycobacterium Tuberculosis, In Lipid Bilayers
Organism: Mycobacterium tuberculosis h37rv
Method: SOLID-STATE NMR Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens, Alphapapillomavirus 10
Method: ELECTRON MICROSCOPY Release Date: 2025-02-26 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Organism: Homo sapiens, Human papillomavirus 11
Method: ELECTRON MICROSCOPY Release Date: 2025-02-26 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Organism: Homo sapiens, Human papillomavirus type 16
Method: ELECTRON MICROSCOPY Release Date: 2025-02-26 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Organism: Homo sapiens, Human papillomavirus 18
Method: ELECTRON MICROSCOPY Release Date: 2025-02-26 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Sars-Cov-2 Envelope Protein Transmembrane Domain: Dimeric Structure Determined By Solid-State Nmr
Organism: Severe acute respiratory syndrome coronavirus 2
Method: SOLID-STATE NMR Release Date: 2023-11-15 Classification: VIRAL PROTEIN |
 |
Crystal Structure Of A Decarboxylase From Trichosporon Moniliiforme In Complex With O-Nitrophenol
Organism: Cutaneotrichosporon moniliiforme
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2023-08-16 Classification: HYDROLASE Ligands: OPO, MG |
 |
Organism: Unclassified marinobacter
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2022-10-19 Classification: HYDROLASE Ligands: ACT, J1K |
 |
Organism: Sinorhizobium fredii ccbau 83666
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2022-08-03 Classification: ISOMERASE Ligands: MG, SO4 |
 |
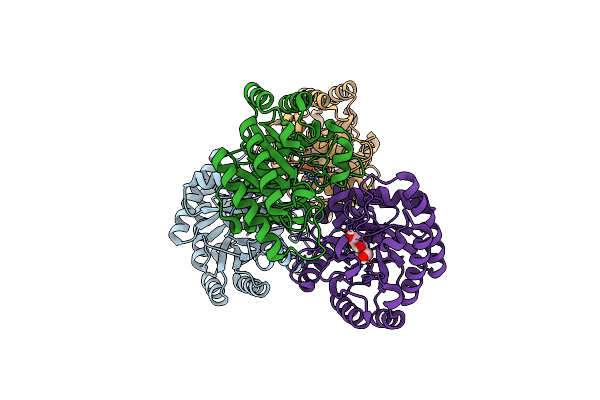
Crystal Structures Of D-Allulose 3-Epimerase With D-Fructose From Sinorhizobium Fredii
Organism: Sinorhizobium fredii ccbau 83666
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2022-08-03 Classification: ISOMERASE Ligands: MG, FUD |
 |
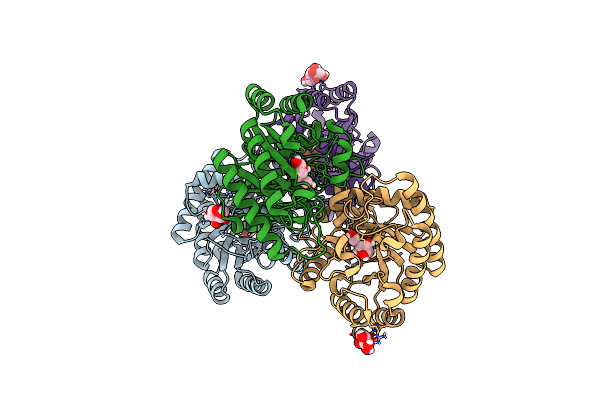
Crystal Structures Of D-Allulose 3-Epimerase With D-Tagatose From Sinorhizobium Fredii
Organism: Sinorhizobium fredii ccbau 83666
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2022-08-03 Classification: ISOMERASE Ligands: MG, TAG |
 |
Crystal Structures Of D-Allulose 3-Epimerase With D-Sorbose From Sinorhizobium Fredii
Organism: Sinorhizobium fredii ccbau 83666
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-08-03 Classification: ISOMERASE Ligands: SDD, HVC, MG |
 |
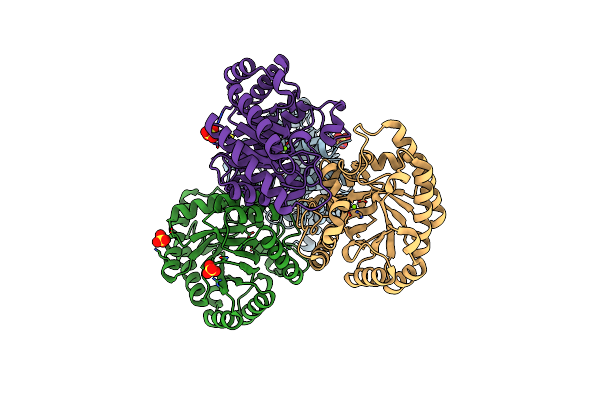
Crystal Structures Of D-Allulose 3-Epimerase With D-Allulose From Sinorhizobium Fredii
Organism: Sinorhizobium fredii ccbau 83666
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-08-03 Classification: ISOMERASE Ligands: MG, PSJ |
 |
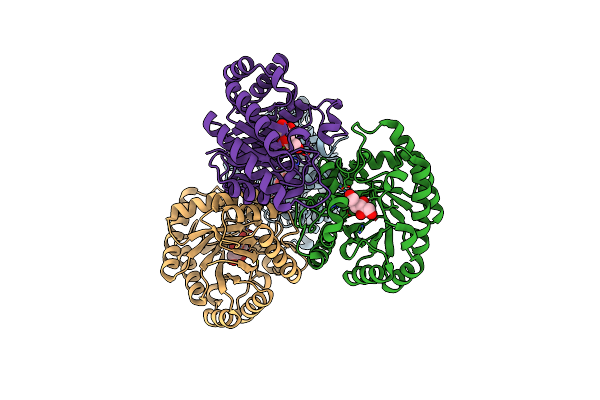
Organism: Agrobacterium sp. sul3
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2022-05-11 Classification: ISOMERASE Ligands: MG, SO4 |
 |
Crystal Structure Of D-Allulose 3-Epimerase With D-Fructose From Agrobacterium Sp. Sul3
Organism: Agrobacterium sp. sul3
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2022-05-11 Classification: ISOMERASE Ligands: FUD, MG |
 |
Crystal Structure Of D-Allulose 3-Epimerase With D-Allulose From Agrobacterium Sp. Sul3
Organism: Agrobacterium sp. sul3
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2022-05-11 Classification: ISOMERASE Ligands: PSJ, MG |
 |
Mouse Toll-Like Receptor 3 Ectodomain In Complex With Lncrna Rmrp In Elongated Form
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2021-10-20 Classification: RNA BINDING PROTEIN/RNA |
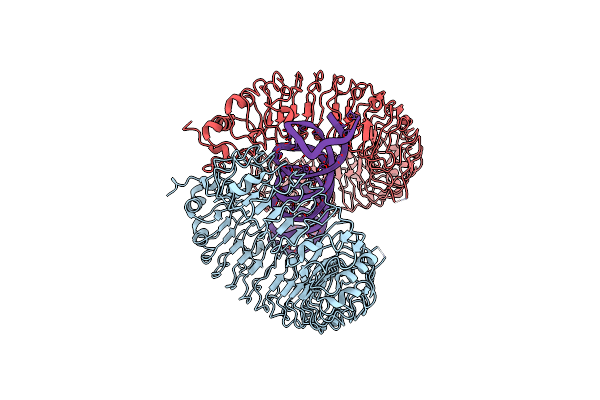 |
Mouse Toll-Like Receptor 3 Ectodomain In Complex With Lncrna Rmrp In Lapped Form
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2021-10-20 Classification: RNA BINDING PROTEIN/RNA |

