Search Count: 144
 |
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: VIRAL PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: RNA BINDING PROTEIN Ligands: ZN |
 |
Electronic Microscopy Structure Of Human Schlafen14-E211A Dimer In Complex With Dsrna
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: RNA BINDING PROTEIN/RNA Ligands: ZN |
 |
Organism: Sphingomonas sp. tpd3009
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: HYDROLASE |
 |
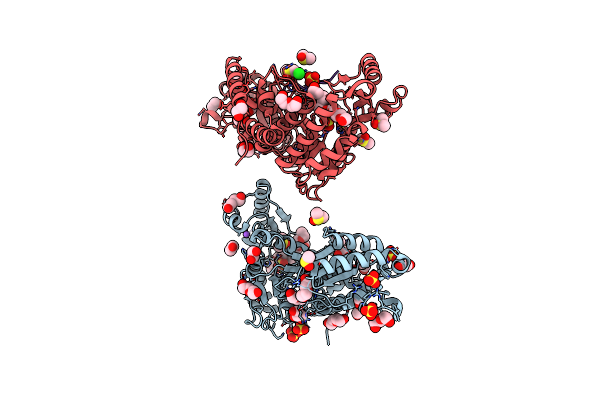
Mycobacterium Tuberculosis Pks13 Acyltransferase Serine Converted To Beta-Lactam Form By Cec215 Via Sufex Reaction
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Release Date: 2025-05-21 Classification: ANTIBIOTIC Ligands: SO4, CL, PG4, EDO, DMS, GOL, PEG |
 |
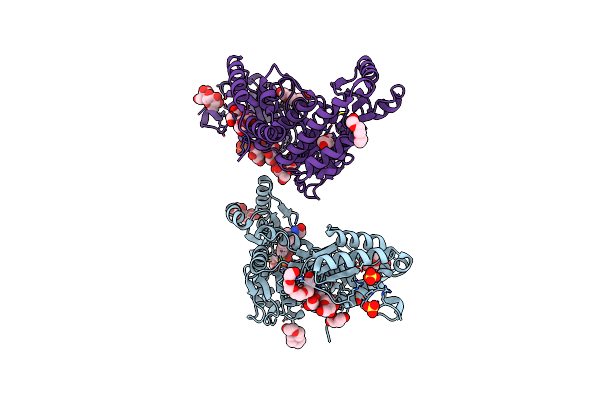
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Release Date: 2025-05-21 Classification: ANTIBIOTIC Ligands: DMS, SO4, PE5, EDO, PEG, GOL, CL, NA, P33 |
 |
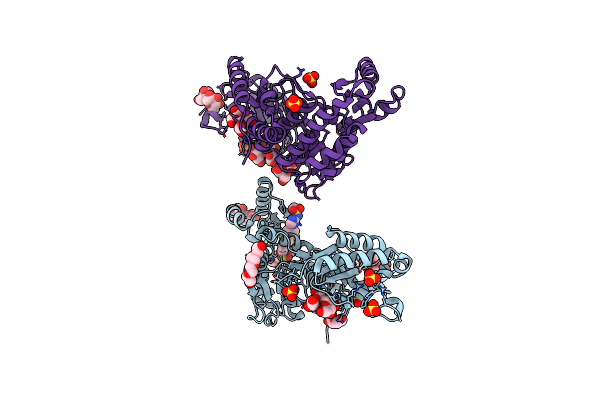
M. Tuberculosis Pks13 Acyltransferase (At) Domain In Complex With Sufex Inhibitor Cec215
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Release Date: 2025-05-07 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 1PE, SO4, A1ATV |
 |
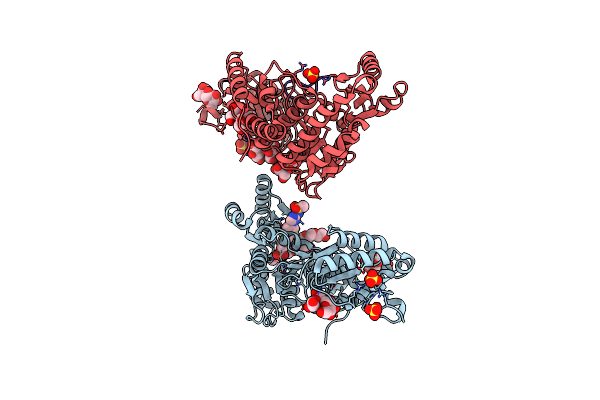
M. Tuberculosis Pks13 Acyltransferase (At) Domain In Complex With Sufex Inhibitor Cmx410
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Release Date: 2025-05-07 Classification: TRANSFERASE/TRANSFERSE INHIBITOR Ligands: 1PE, SO4, A1ATW |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Release Date: 2025-05-07 Classification: TRANSFERASE Ligands: 1PE, SO4 |
 |
M. Tuberculosis Pks13 Acyltransferase (At) Domain In Complex With Sufex Inhibitor Cmx410 - Reaction Product
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Release Date: 2025-05-07 Classification: TRANSFERASE Ligands: SO4, 1PE, A1AVL |
 |
Organism: Bacillus subtilis a29
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: HYDROLASE |
 |
Organism: Bacillus subtilis a29
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: HYDROLASE |
 |
Organism: Bacillus subtilis a29
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: HYDROLASE |
 |
Organism: Bacillus subtilis a29
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: HYDROLASE |
 |
Organism: Bacillus subtilis a29
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: HYDROLASE |
 |
Organism: Bacillus subtilis a29
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: HYDROLASE |
 |
Organism: Bacillus subtilis a29
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: HYDROLASE |
 |
Organism: Bacillus subtilis a29
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: HYDROLASE |
 |
Organism: Bacillus subtilis a29
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: HYDROLASE |
 |
Organism: Isochrysis galbana
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: PHOTOSYNTHESIS Ligands: CLA, KC2, DD6, LHG, LMG, A86, LMU, SQD, DGD, BCR, SF4, PQN |

