Search Count: 30
 |
Cryo-Em Structure Of Rhesus Antibody T646-A.01 In Complex With Hiv-1 Env Trimer Q23.17 Md39
Organism: Human immunodeficiency virus 1, Macaca mulatta
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Macaca mulatta, Human immunodeficiency virus 1
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Human immunodeficiency virus 1, Macaca mulatta
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Macaca mulatta, Human immunodeficiency virus 1
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Cryo-Em Structure Of Rhesus Antibody 41328-A.01 In Complex With Hiv-1 Env Bg505 Ds-Sosip
Organism: Human immunodeficiency virus 1, Macaca mulatta
Method: ELECTRON MICROSCOPY Release Date: 2025-05-21 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Cryo-Em Structure Of Rhesus Antibody V031-A.01 In Complex With Hiv-1 Env Bg505 Ds-Sosip
Organism: Macaca mulatta, Human immunodeficiency virus 1
Method: ELECTRON MICROSCOPY Release Date: 2025-05-07 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Cryo-Em Structure Of Rhesus Antibody 6070-A.01 In Complex With Hiv-1 Env Trimer Q23.17 Md39
Organism: Human immunodeficiency virus 1, Macaca mulatta
Method: ELECTRON MICROSCOPY Release Date: 2025-05-07 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Cryo-Em Structure Of Rhesus Antibody 44715-A.01 In Complex With Hiv-1 Env Bg505 Ds-Sosip
Organism: Human immunodeficiency virus 1, Macaca mulatta
Method: ELECTRON MICROSCOPY Release Date: 2025-05-07 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Cryo-Em Structure Of Rhesus Antibody V033-A.01 In Complex With Hiv-1 Env Bg505 Ds-Sosip
Organism: Human immunodeficiency virus 1, Macaca mulatta
Method: ELECTRON MICROSCOPY Release Date: 2025-05-07 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-05-07 Classification: PROTEIN FIBRIL |
 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-05-07 Classification: PROTEIN FIBRIL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-04-16 Classification: IMMUNE SYSTEM Ligands: CL, CA, Y4T |
 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: PROTEIN FIBRIL Ligands: OG9 |
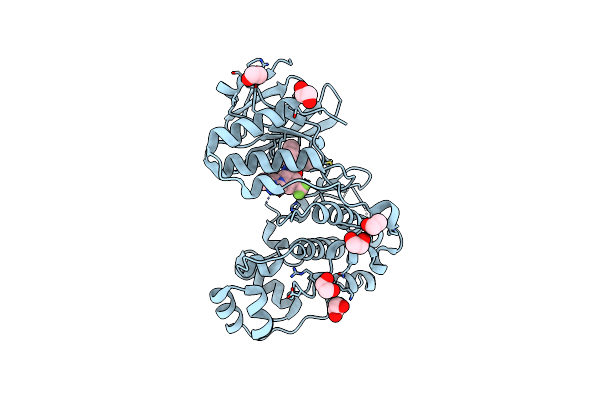 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-11-13 Classification: TRANSFERASE Ligands: EDO, NIL |
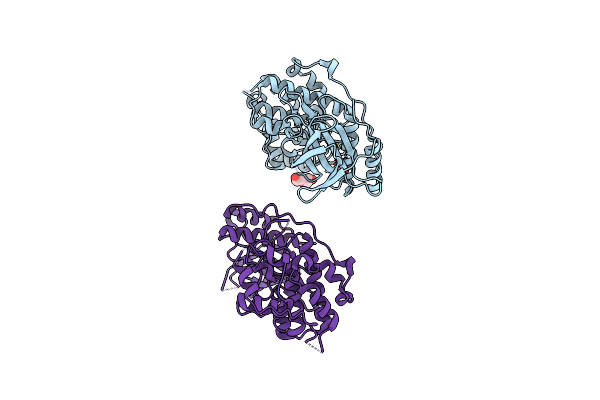 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.83 Å Release Date: 2024-11-13 Classification: TRANSFERASE Ligands: PEG, EDO |
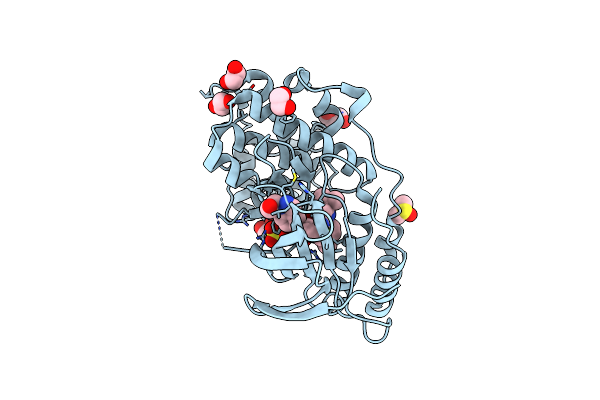 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-11-13 Classification: TRANSFERASE Ligands: SO4, EDO, GOL, DMS, PDO, A1AWV |
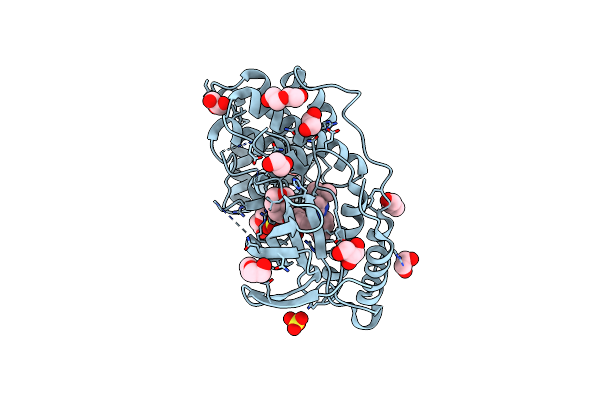 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-11-13 Classification: TRANSFERASE Ligands: B96, SO4, EDO, PEG, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2024-11-13 Classification: TRANSFERASE Ligands: EDO, A1AWV, PDO |
 |
Organism: Anaeroglobus
Method: ELECTRON MICROSCOPY Release Date: 2024-09-04 Classification: DNA BINDING PROTEIN/RNA Ligands: MG |
 |
Solution Structure Of The Periplasmic Domain Of The Anti-Sigma Factor Rsgi1 From Clostridium Thermocellum
Organism: Acetivibrio thermocellus dsm 1313
Method: SOLUTION NMR Release Date: 2023-05-24 Classification: SIGNALING PROTEIN |

