Search Count: 41
 |
Organism: Enterovirus d68
Method: ELECTRON MICROSCOPY Release Date: 2025-03-26 Classification: VIRUS |
 |
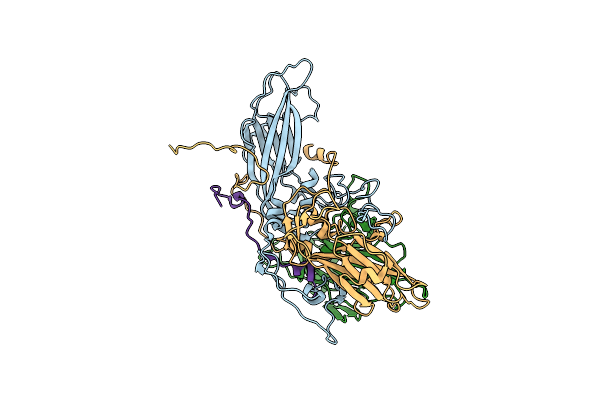
Cryo-Em Structure Of Human Enterovirus D68 Usa/Il/14-18952 In Complex With Fc-Mfsd6(L3)
Organism: Homo sapiens, Enterovirus d68
Method: ELECTRON MICROSCOPY Release Date: 2025-03-26 Classification: VIRUS |
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-01-08 Classification: TRANSPORT PROTEIN Ligands: Z41, PLM |
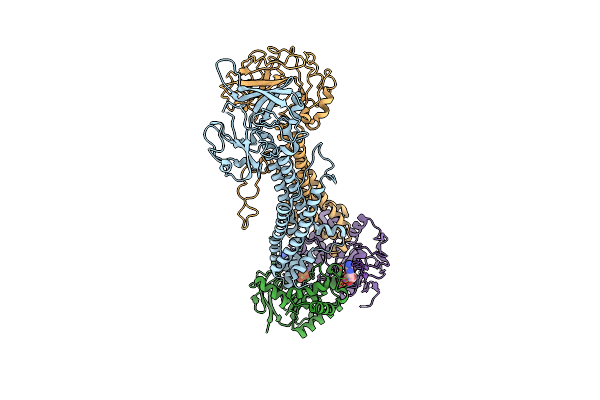 |
Cryo-Em Structure Of Endogenous Atp-Bound Lolcde With Lold-E171Q Mutations In Nanodiscs
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-01-08 Classification: TRANSPORT PROTEIN Ligands: MG, ATP |
 |
Organism: Nicotiana benthamiana
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: PLANT PROTEIN Ligands: ATP |
 |
Organism: Nicotiana benthamiana
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: PLANT PROTEIN Ligands: ATP |
 |
Cryo-Em Structure Of Expanded Coxsackievirus A16 Empty Particle After Incubation With 8C4 Antibody
Organism: Coxsackievirus a16
Method: ELECTRON MICROSCOPY Release Date: 2022-10-19 Classification: STRUCTURAL PROTEIN |
 |
Cryo-Em Structure Of Expanded Coxsackievirus A16 Empty Particle In Complex With Antibody 9B5
Organism: Coxsackievirus a16
Method: ELECTRON MICROSCOPY Release Date: 2022-10-19 Classification: STRUCTURAL PROTEIN |
 |
Cryo-Em Structure Of Compact Ca16 Empty Particle In Complex With A Neutralizing Antibody 8C4
Organism: Coxsackievirus a16
Method: ELECTRON MICROSCOPY Release Date: 2022-09-14 Classification: STRUCTURAL PROTEIN Ligands: SPH |
 |
Cryo-Em Structure Of Compact Coxsackievirus A16 Empty Particle In Complex With A Neutralizing Antibody 9B5
Organism: Coxsackievirus a16
Method: ELECTRON MICROSCOPY Release Date: 2022-09-14 Classification: STRUCTURAL PROTEIN Ligands: SPH |
 |
Organism: Coxsackievirus a16
Method: ELECTRON MICROSCOPY Release Date: 2022-08-24 Classification: VIRUS Ligands: SPH |
 |
Cryo-Em Structure Of Coxsackievirus A16 In Complex With A Neutralizing Antibody 9B5
Organism: Coxsackievirus a16
Method: ELECTRON MICROSCOPY Release Date: 2022-08-24 Classification: VIRUS |
 |
Organism: Escherichia coli (strain k12), Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2021-04-07 Classification: PROTEIN TRANSPORT Ligands: Z41, PLM |
 |
Organism: Escherichia coli (strain k12)
Method: ELECTRON MICROSCOPY Release Date: 2021-04-07 Classification: PROTEIN TRANSPORT |
 |
Organism: Escherichia coli (strain k12), Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2021-04-07 Classification: PROTEIN TRANSPORT Ligands: ANP, MG, Z41, PLM |
 |
Organism: Escherichia coli (strain k12)
Method: ELECTRON MICROSCOPY Release Date: 2021-04-07 Classification: PROTEIN TRANSPORT Ligands: ANP, MG |
 |
Organism: Escherichia coli (strain k12), Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2021-04-07 Classification: PROTEIN TRANSPORT Ligands: Z41, ADP, MG, PLM |
 |
Organism: Escherichia coli (strain k12), Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2021-04-07 Classification: PROTEIN TRANSPORT Ligands: Z41, PLM |
 |
S-3C1-F3B Structure, All The Three Rbds Are In The Up Conformation And Each Of Them Associates With A 3C1 Fab
Organism: Severe acute respiratory syndrome coronavirus 2, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2020-12-02 Classification: VIRAL PROTEIN |
 |
S-3C1-F3A Structure, Two Rbds Are Up And One Rbd Is Down, Each Rbd Binds With A 3C1 Fab.
Organism: Severe acute respiratory syndrome coronavirus 2, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2020-12-02 Classification: VIRAL PROTEIN |

