Search Count: 54
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: TRANSPORT PROTEIN |
 |
Structure Of Arabidopsis Thaliana Abcb1 With Amp-Pnp Bound In The Inward-Facing Conformation
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: TRANSPORT PROTEIN Ligands: MG, ANP |
 |
Structure Of Arabidopsis Thaliana Abcb1 With Brassinolide Bound In The Inward-Facing Conformation
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Resolution:3.10 Å Release Date: 2025-03-05 Classification: TRANSPORT PROTEIN Ligands: BLD |
 |
Structure Of Arabidopsis Thaliana Abcb1 With Brassinolide And Amp-Pnp Bound In The Inward-Facing Conformation
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Resolution:3.10 Å Release Date: 2025-03-05 Classification: TRANSPORT PROTEIN Ligands: MG, ANP, BLD |
 |
Structure Of Arabidopsis Thaliana Abcb1 In The Inward-Facing Conformation Under Iaa Condition
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: TRANSPORT PROTEIN |
 |
Structure Of Arabidopsis Thaliana Abcb1 With Amp-Pnp Bound In The Inward-Facing Conformation Under Iaa Condition
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: TRANSPORT PROTEIN Ligands: MG, ANP |
 |
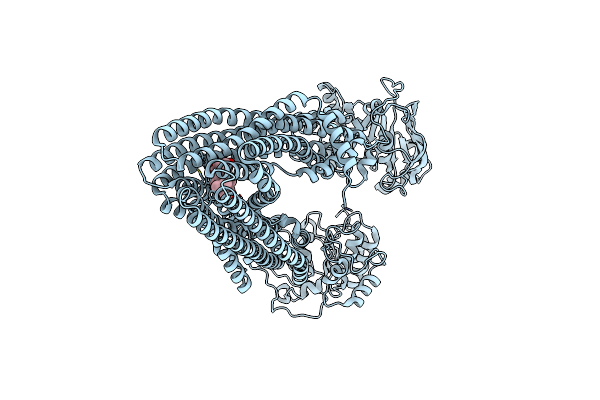
Structure Of Arabidopsis Thaliana Abcb1 With Amp-Pnp Bound In The Outward-Facing Conformation
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Resolution:3.10 Å Release Date: 2025-03-05 Classification: TRANSPORT PROTEIN Ligands: MG, ANP |
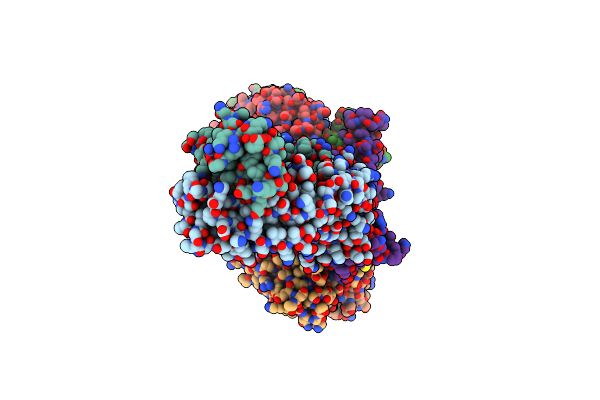 |
Cryo-Em Structure Of Sars-Cov-2 Xbb.1.5 Receptor-Binding Domain (Rbd) Complexed With Cb6 Mutant,S309, And S304 Antibodies
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2024-10-16 Classification: IMMUNE SYSTEM/VIRAL PROTEIN |
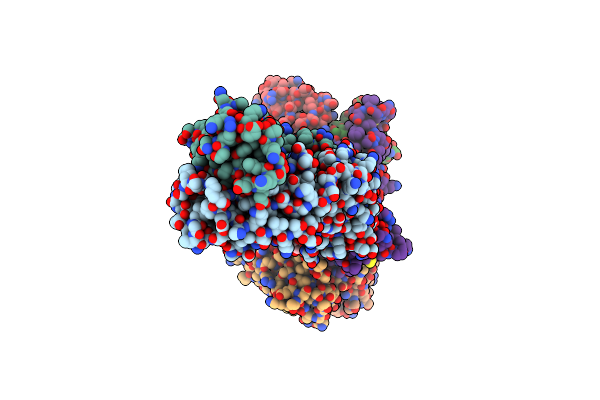 |
Cryo-Em Structure Of Sars-Cov-2 Receptor-Binding Domain (Rbd) Complexed With Cb6 Mutant,S309, And S304 Antibodies
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2024-10-16 Classification: IMMUNE SYSTEM/VIRAL PROTEIN |
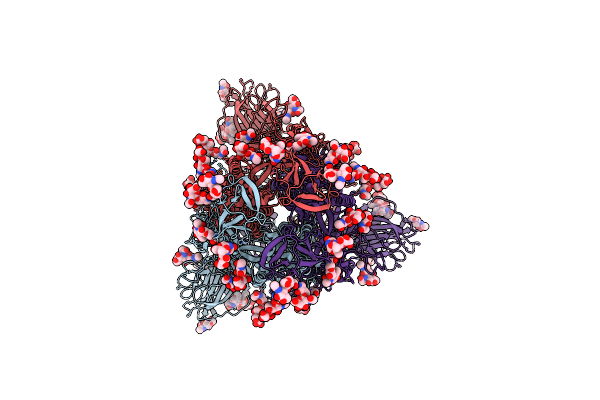 |
Organism: Bat sars-like coronavirus rsshc014, Enterobacteria phage t6
Method: ELECTRON MICROSCOPY Release Date: 2024-07-17 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Bat sars-like coronavirus wiv1, Enterobacteria phage t6
Method: ELECTRON MICROSCOPY Release Date: 2024-07-17 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Bat sars-like coronavirus wiv1, Enterobacteria phage t6, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-07-17 Classification: VIRAL PROTEIN/HYDROLASE Ligands: NAG |
 |
Organism: Bat sars-like coronavirus wiv1
Method: ELECTRON MICROSCOPY Release Date: 2024-07-17 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2024-02-14 Classification: TRANSPORT PROTEIN |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2024-02-14 Classification: TRANSPORT PROTEIN Ligands: K |
 |
Organism: Triticum aestivum
Method: ELECTRON MICROSCOPY Release Date: 2024-02-14 Classification: TRANSPORT PROTEIN |
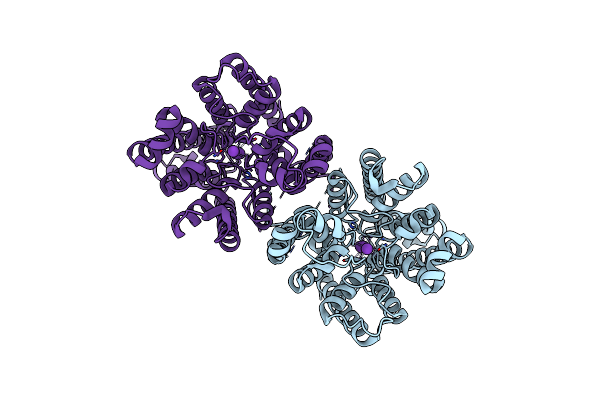 |
Organism: Triticum aestivum
Method: ELECTRON MICROSCOPY Release Date: 2024-02-14 Classification: TRANSPORT PROTEIN Ligands: K |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2024-01-17 Classification: TRANSPORT PROTEIN |
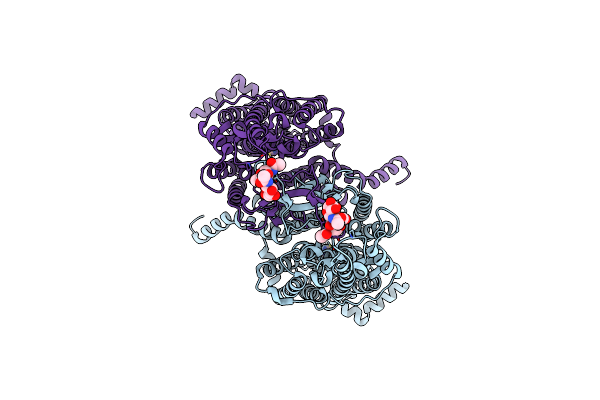 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2024-01-17 Classification: TRANSPORT PROTEIN Ligands: ADE |
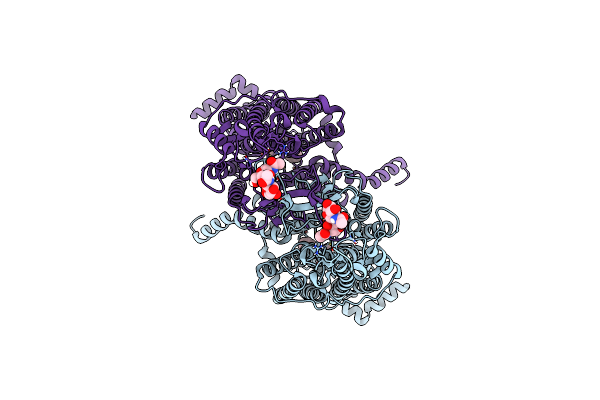 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2024-01-17 Classification: TRANSPORT PROTEIN Ligands: EMU |

