Search Count: 1,209
All
Selected
 |
Glutamate Transporter Homologue Gltph Mutant P206R In Complex With L-Aspartate And Sodium Ions In Salipro
Organism: Pyrococcus horikoshii
Method: ELECTRON MICROSCOPY Resolution:3.34 Å Release Date: 2026-02-18 Classification: MEMBRANE PROTEIN Ligands: ASP, NA, PG8, PGM, CL, D3D |
 |
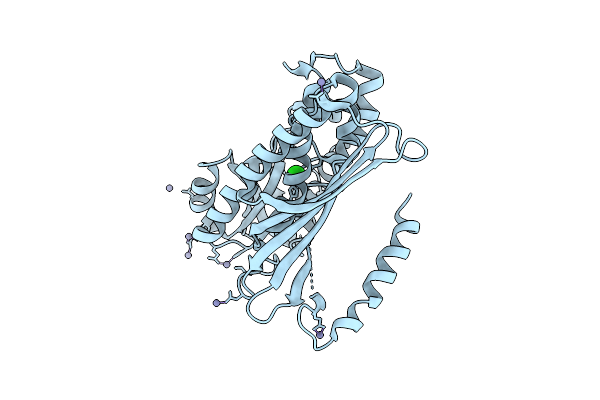
Organism: Pyrococcus horikoshii ot3
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2025-11-05 Classification: METAL BINDING PROTEIN Ligands: NI, NAD, 1BO |
 |
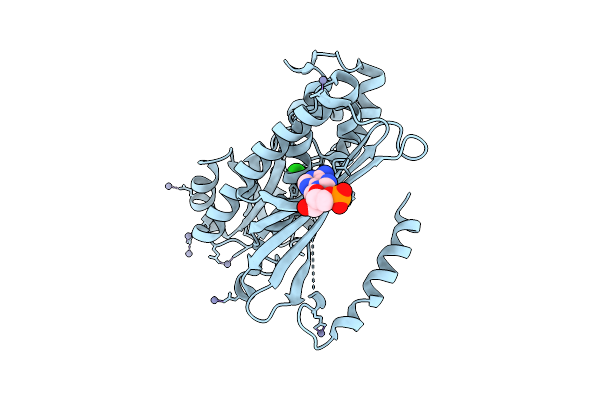
Organism: Pyrococcus horikoshii ot3
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2025-10-15 Classification: LIGASE Ligands: ZN, CL |
 |
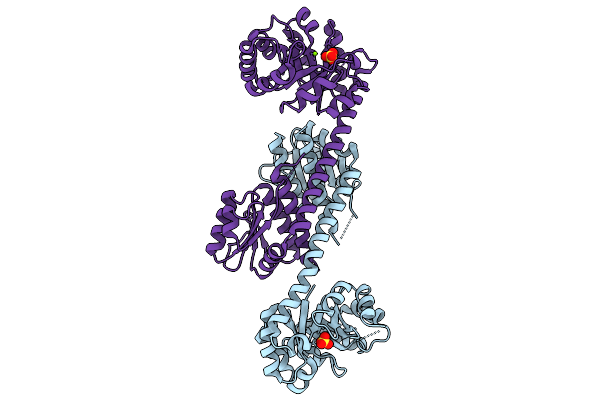
Organism: Pyrococcus horikoshii ot3
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2025-10-15 Classification: LIGASE Ligands: AMP, ZN, CL |
 |
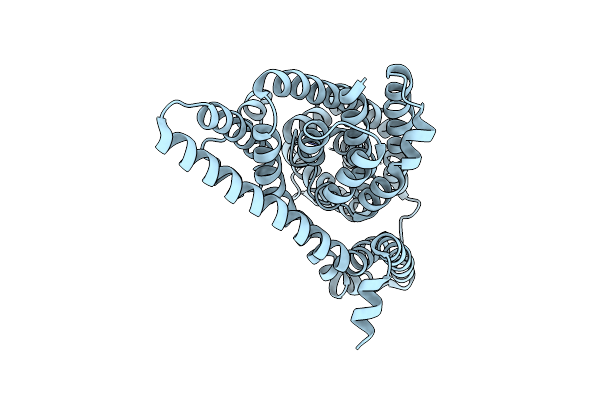
Crystal Structure Of A Bifunctional 3-Hexulose-6-Phosphate Synthase/6-Phospho-3-Hexuloisomerase
Organism: Pyrococcus horikoshii ot3
Method: X-RAY DIFFRACTION Resolution:2.64 Å Release Date: 2025-10-01 Classification: ISOMERASE Ligands: MG, SO4 |
 |
Organism: Pyrococcus horikoshii
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: TRANSPORT PROTEIN |
 |
Organism: Pyrococcus horikoshii
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: TRANSPORT PROTEIN |
 |
Organism: Pyrococcus horikoshii ot3
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2025-03-12 Classification: TRANSFERASE Ligands: SO4, SO3, EDO, PEG |
 |
Crystal Structure Of Nep1 In Complex With Adenosine From Pyrococcus Horikoshii Ot3
Organism: Pyrococcus horikoshii ot3
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2025-03-12 Classification: TRANSFERASE Ligands: CL, ADN, SO4, SO3, EDO, PEG |
 |
Crystal Structure Of Nep1 In Complex With 5'-Methylthioadenosine From Pyrococcus Horikoshii Ot3
Organism: Pyrococcus horikoshii ot3
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-03-12 Classification: TRANSFERASE Ligands: CL, MTA, EDO, GOL, ACT |
 |
Crystal Structure Of Udp-Galactose 4-Epimerase From Pyrococcus Horikoshii With Bound Nad
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-12-18 Classification: ISOMERASE Ligands: NAD |
 |
Crystal Structure Of Udp-Galactose 4-Epimerase From Pyrococcus Horikoshii With Bound Nad And Gdp-L-Fucose
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2024-12-18 Classification: ISOMERASE Ligands: NAD, GFB |
 |
Crystal Structure Of Udp-Galactose 4-Epimerase From Pyrococcus Horikoshii Containing Y145F Mutation And With Bound Nad And Gdp-L-Fucose
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2024-12-18 Classification: ISOMERASE Ligands: NAD, GFB |
 |
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:2.94 Å Release Date: 2024-10-30 Classification: METAL BINDING PROTEIN Ligands: ATP, MG |
 |
Crystal Structure Of Rrna (Uracil-C5)-Methyltransferase From Pyrococcus Horikoshii Ot3
Organism: Pyrococcus horikoshii ot3
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2024-10-16 Classification: TRANSFERASE |
 |
Crystal Structure Of Dimt1 From The Thermophilic Archaeon, Pyrococcus Horikoshii
Organism: Pyrococcus horikoshii ot3
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2024-08-28 Classification: TRANSFERASE Ligands: ZN, SO3, GOL, PEG, EDO, ACT, ARG, EPE |
 |
Crystal Structure Of Dimt1 In Complex With 5'-Methylthioadenosine And Adenosine From Pyrococcus Horikoshii
Organism: Pyrococcus horikoshii ot3
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2024-08-28 Classification: TRANSFERASE Ligands: MTA, ADN, ZN, CL, SO3, GOL, EDO, ACT, ARG, PEG |
 |
Crystal Structure Of Dimt1 In Complex With 5'-Methylthioadenosine From Pyrococcus Horikoshii (Formi)
Organism: Pyrococcus horikoshii ot3
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-08-28 Classification: TRANSFERASE Ligands: MTA, ZN, SO3, PEG, CL, GOL, EDO |
 |
Crystal Structure Of Dimt1 In Complex With 5'-Methylthioadenosine From Pyrococcus Horikoshii (Formii)
Organism: Pyrococcus horikoshii ot3
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-08-28 Classification: TRANSFERASE Ligands: MTA, ZN, SO3, GOL, EDO, PEG, PGE, ACT |
 |
Crystal Structure Of Dimt1 In Complex With Adenosylornithine (Sfg) From Pyrococcus Horikoshii
Organism: Pyrococcus horikoshii ot3
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-08-28 Classification: TRANSFERASE Ligands: SFG, ZN, CL, PEG, ARG, SO3, EDO |

