Search Count: 1,100
All
Selected
 |
Crystal Structure Of S-Adenosyl-L-Homocysteine Hydrolase From Pyrococcus Furiosus In Complex With Hypoxanthine
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2026-03-04 Classification: HYDROLASE Ligands: NAD, HPA |
 |
Crystal Structure Of S-Adenosyl-L-Homocysteine Hydrolase Mutant C221W From Pyrococcus Furiosus In Complex With Inosine
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.53 Å Release Date: 2026-03-04 Classification: HYDROLASE Ligands: NOS |
 |
Organism: Archaeoglobus fulgidus dsm 4304, Pyrococcus furiosus dsm 3638
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: ANTIVIRAL PROTEIN Ligands: MG, ATP, ZN |
 |
Organism: Archaeoglobus fulgidus dsm 4304, Archaeoglobus fulgidus, Pyrococcus furiosus dsm 3638
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: ANTIVIRAL PROTEIN Ligands: MG, ATP, ZN |
 |
Crystal Structure Of Proline Dipeptidase From Pyrococcus Furiosus Complexed With Co
Organism: Pyrococcus furiosus dsm 3638
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-12-10 Classification: HYDROLASE |
 |
Crystal Structure Of Proline Dipeptidase From Pyrococcus Furiosus Complexed With Mn
Organism: Pyrococcus furiosus dsm 3638
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2025-12-10 Classification: HYDROLASE Ligands: MN, NA, PEG |
 |
Organism: Archaeoglobus fulgidus dsm 4304, Pyrococcus furiosus dsm 3638, Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:3.40 Å Release Date: 2025-12-10 Classification: RNA BINDING PROTEIN/RNA/DNA |
 |
Crystal Structure Of Upf0235 Protein Pf1765 From Pyrococcus Furiosus Crystallized With Sodium Acetate At Ph 8
Organism: Pyrococcus furiosus dsm 3638
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2025-11-26 Classification: UNKNOWN FUNCTION |
 |
Crystal Structure Of Upf0235 Protein Pf1765 From Pyrococcus Furiosus At Ph 6
Organism: Pyrococcus furiosus dsm 3638
Method: X-RAY DIFFRACTION Resolution:1.13 Å Release Date: 2025-11-26 Classification: UNKNOWN FUNCTION |
 |
Crystal Structure Of Upf0235 Protein Pf1765 From Pyrococcus Furiosus At Ph 9
Organism: Pyrococcus furiosus dsm 3638
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2025-11-26 Classification: UNKNOWN FUNCTION |
 |
Crystal Structure Of Upf0235 Protein Pf1765 From Pyrococcus Furiosus Crystallized With Sodium Citrate At Ph 8.0
Organism: Pyrococcus furiosus dsm 3638
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2025-11-26 Classification: UNKNOWN FUNCTION |
 |
Crystal Structure Of Upf0235 Protein Pf1765 From Pyrococcus Furiosus Crystallized With Peg1500 And Glycerol At Ph 8.0
Organism: Pyrococcus furiosus dsm 3638
Method: X-RAY DIFFRACTION Resolution:1.11 Å Release Date: 2025-11-26 Classification: UNKNOWN FUNCTION Ligands: GOL, CL |
 |
Crystal Structure Of Upf0235 Protein Pf1765 From Pyrococcus Furiosus At Ph 4
Organism: Pyrococcus furiosus dsm 3638
Method: X-RAY DIFFRACTION Resolution:1.19 Å Release Date: 2025-11-26 Classification: UNKNOWN FUNCTION |
 |
Crystal Structure Of Upf0235 Protein Pf1765 From Pyrococcus Furiosus Crystallized With Cacl2 At Ph 5
Organism: Pyrococcus furiosus dsm 3638
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-11-26 Classification: UNKNOWN FUNCTION |
 |
Crystal Structure Of Upf0235 Protein Pf1765 From Pyrococcus Furiosus Crystallized With Cacl2 At Ph 6
Organism: Pyrococcus furiosus dsm 3638
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-11-26 Classification: UNKNOWN FUNCTION Ligands: CL, NA |
 |
Crystal Structure Of Upf0235 Protein Pf1765 From Pyrococcus Furiosus Crystallized With Cacl2 At Ph 7
Organism: Pyrococcus furiosus dsm 3638
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2025-11-26 Classification: UNKNOWN FUNCTION |
 |
Organism: Pyrococcus furiosus dsm 3638
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-11-19 Classification: METAL BINDING PROTEIN |
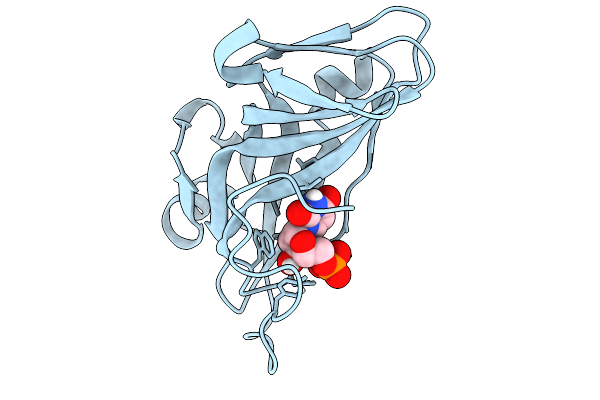 |
Organism: Pyrococcus furiosus dsm 3638
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-11-19 Classification: METAL BINDING PROTEIN Ligands: UMP |
 |
Organism: Pyrococcus furiosus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: RIBOSOME Ligands: ZN |
 |
Organism: Pyrococcus furiosus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: RIBOSOME Ligands: ZN |

