Search Count: 6
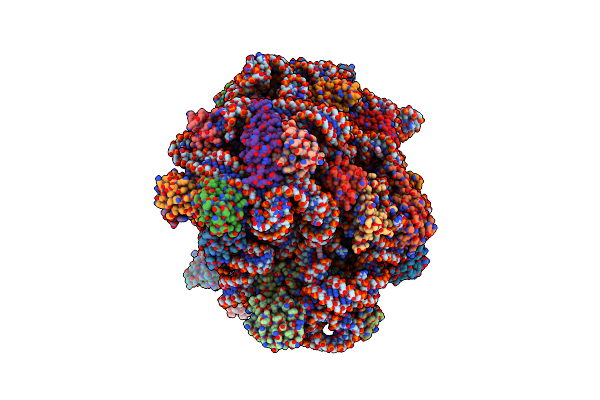 |
Cryo-Em Structure Of The Pyrobaculum Calidifontis 50S Ribosomal Subunit In Complex With Dri
Organism: Pyrobaculum calidifontis jcm 11548
Method: ELECTRON MICROSCOPY Release Date: 2024-11-20 Classification: RIBOSOME Ligands: SPM, MG, ZN |
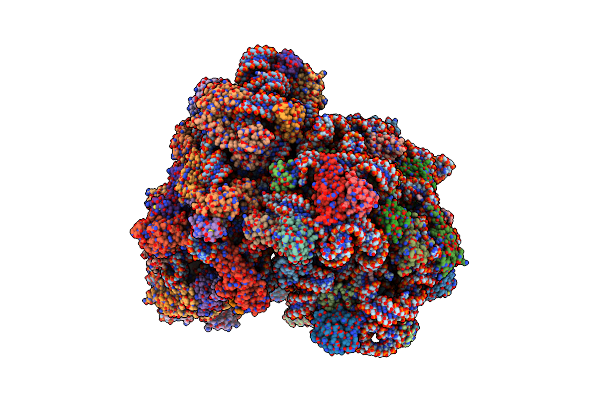 |
Organism: Pyrobaculum calidifontis jcm 11548
Method: ELECTRON MICROSCOPY Release Date: 2024-11-20 Classification: RIBOSOME Ligands: SPM, MG, ZN |
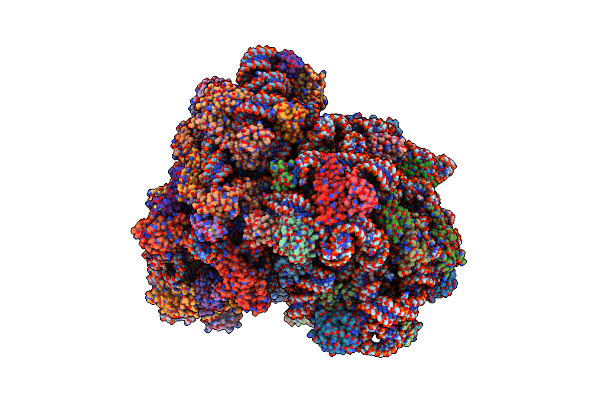 |
Cryo-Em Structure Of The Pyrobaculum Calidifontis 70S Ribosome In Complex With Dri
Organism: Pyrobaculum calidifontis jcm 11548
Method: ELECTRON MICROSCOPY Release Date: 2024-11-20 Classification: RIBOSOME Ligands: SPM, MG, ZN |
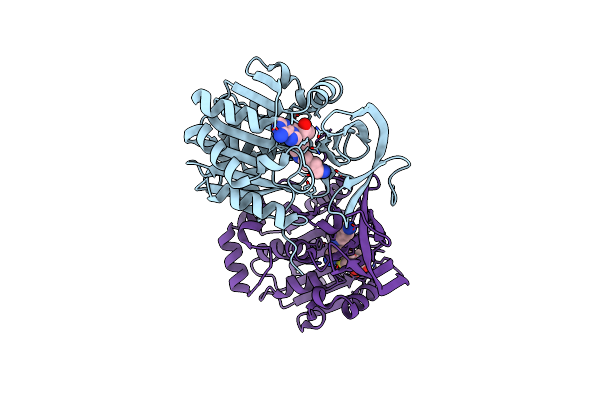 |
Crystal Structure Of The Aminopropyltransferase, Spee From Hyperthermophilic Crenarchaeon, Pyrobaculum Calidifontis In Complex With 5'-Methylthioadenosine (Mta) And Spermidine
Organism: Pyrobaculum calidifontis jcm 11548
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2022-06-15 Classification: TRANSFERASE Ligands: MTA, SPD |
 |
Organism: Pyrobaculum calidifontis jcm 11548
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2016-10-12 Classification: OXIDOREDUCTASE Ligands: ZN, SO4, PPV, NDP |
 |
Crystal Structure And Nuclease Activity Of The Crispr-Associated Cas4 Protein Pcal_0546 From Pyrobaculum Calidifontis Containing A [2Fe-2S] Cluster
Organism: Pyrobaculum calidifontis jcm 11548
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2014-09-17 Classification: HYDROLASE Ligands: MG, FES |

