Search Count: 17
 |
Nmr Solution Structure Of The Sylf Domain Of Burkholderia Pseudomallei Bpsl1445
Organism: Burkholderia pseudomallei
Method: SOLUTION NMR Release Date: 2021-12-29 Classification: LIPID BINDING PROTEIN |
 |
Organism: Pseudomonas denitrificans (nomen rejiciendum)
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2018-05-23 Classification: TRANSPORT PROTEIN Ligands: BOG |
 |
Organism: Pseudomonas fragi a22
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2018-05-16 Classification: TRANSPORT PROTEIN |
 |
Crystal Structure Of The Transporter Chbc, The Iic Component From The N,N'-Diacetylchitobiose-Specific Phosphotransferase System
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2011-04-06 Classification: membrane protein, transport protein Ligands: ZDM, CIT |
 |
Crystal Structure Of The Potassium Transporter Trkh From Vibrio Parahaemolyticus
Organism: Vibrio parahaemolyticus
Method: X-RAY DIFFRACTION Resolution:3.51 Å Release Date: 2011-01-19 Classification: TRANSPORT PROTEIN Ligands: K |
 |
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2010-05-12 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION Ligands: BOG |
 |
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2010-05-12 Classification: Structural Genomics, Unknown function Ligands: BOG |
 |
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2010-05-12 Classification: Structural Genomics, Unknown function Ligands: BOG |
 |
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2010-05-12 Classification: Structural Genomics, Unknown function Ligands: BOG |
 |
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2010-05-12 Classification: Structural Genomics, Unknown function Ligands: BOG |
 |
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2010-05-12 Classification: Structural Genomics, Unknown function Ligands: BOG |
 |
Solution Nmr Structure Of Protein Yiis From Shigella Flexneri. Northeast Structural Genomics Consortium Target Sfr90
Organism: Shigella flexneri 2a
Method: SOLUTION NMR Release Date: 2008-07-08 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION |
 |
Nmr Solution Structure Of Protein Vpa0419 From Vibrio Parahaemolyticus. Northeast Structural Genomics Target Vpr68
Organism: Vibrio parahaemolyticus rimd 2210633
Method: SOLUTION NMR Release Date: 2008-02-19 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION |
 |
Nmr Structure Of Rpa0253 From Rhodopseudomonas Palustris. Northeast Structural Genomics Consortium Target Rpr3
Organism: Rhodopseudomonas palustris
Method: SOLUTION NMR Release Date: 2006-04-11 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION |
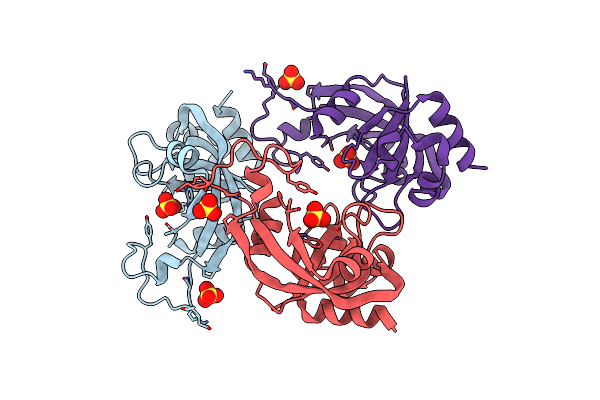 |
X-Ray Crystal Structure Protein Q88Ch6 From Pseudomonas Putida. Northeast Structural Genomics Consortium Target Ppr72.
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2006-03-14 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION Ligands: SO4 |
 |
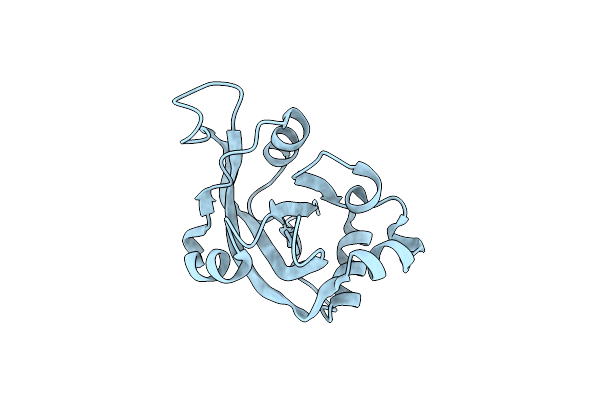
X-Ray Crystal Structure Of Protein Pspto5229 From Pseudomonas Syringae. Northeast Structural Genomics Consortium Target Psr62
Organism: Pseudomonas syringae pv. tomato str. dc3000
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2005-11-08 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION Ligands: MPO, EDO, 144 |
 |
X-Ray Crystal Structure Of Protein Atu2648 From Agrobacterium Tumefaciens. Northeast Structural Genomics Consortium Target Atr33.
Organism: Agrobacterium tumefaciens str. c58
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2005-04-19 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION |

