Search Count: 19
 |
Identification And Optimisation Of Novel Inhibitors Of The Polyketide Synthetase 13 Thioesterase Domain With Antitubercular Activity
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-11-22 Classification: TRANSFERASE Ligands: IJJ |
 |
Identification And Optimisation Of Novel Inhibitors Of The Polyketide Synthetase 13 Thioesterase Domain With Antitubercular Activity
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-11-22 Classification: TRANSFERASE Ligands: SO4, IKG |
 |
Identification And Optimisation Of Novel Inhibitors Of The Polyketide Synthetase 13 Thioesterase Domain With Antitubercular Activity
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2023-11-22 Classification: TRANSFERASE Ligands: SO4, IL8, GOL, 2Q5 |
 |
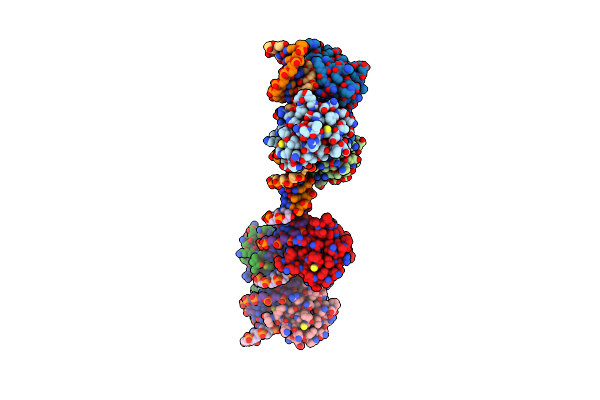
Crystal Structure Of Hsf1 Dna-Binding Domain In Complex With 2-Site Hse Dna In The Head-To-Head Orientation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-07-14 Classification: DNA BINDING PROTEIN/DNA |
 |
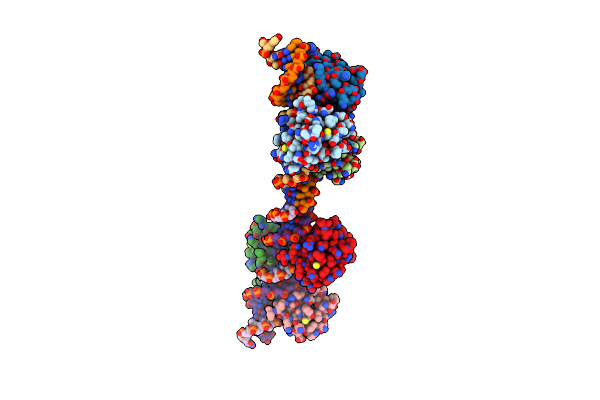
Crystal Structure Of Hsf1 Dna-Binding Domain In Complex With 3-Site Hse Dna (23 Bp)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2021-07-14 Classification: DNA BINDING PROTEIN/DNA |
 |
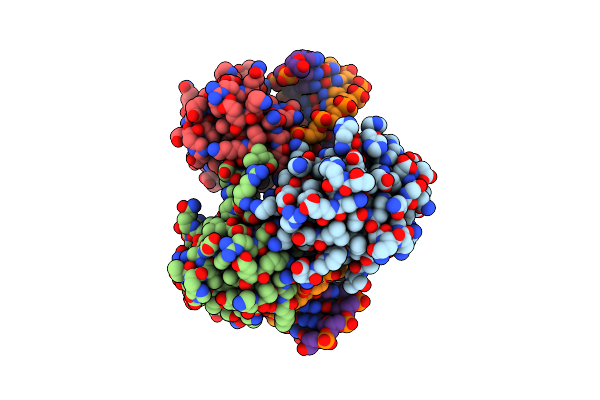
Crystal Structure Of Hsf1 Dna-Binding Domain In Complex With 3-Site Hse Dna (24 Bp)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2021-07-14 Classification: DNA BINDING PROTEIN/DNA |
 |
Crystal Structure Of Hsf2 Dna-Binding Domain In Complex With 3-Site Hse Dna (21 Bp)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2021-07-14 Classification: DNA BINDING PROTEIN/DNA |
 |
Crystal Structure Of E. Coli Seryl-Trna Synthetase Complexed To Seryl Sulfamoyl Adenosine
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-01-22 Classification: LIGASE Ligands: SSA, PO4, EDO |
 |
Crystal Structure Of S. Aureus Seryl-Trna Synthetase Complexed To Seryl Sulfamoyl Adenosine
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2020-01-22 Classification: LIGASE Ligands: SSA, EDO, PEG, MG |
 |
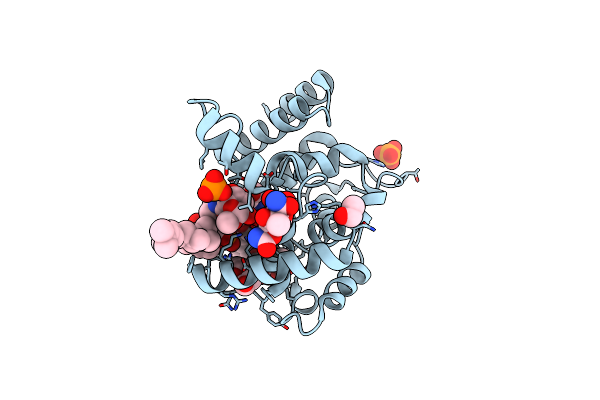
Crystal Structure Of E. Coli Seryl-Trna Synthetase Complexed To A Seryl Sulfamoyl Adenosine Derivative
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2020-01-22 Classification: LIGASE Ligands: JPE, CL, SO4, DMS, EDO, TRS |
 |
Staphylococcus Aureus Monofunctional Glycosyltransferase In Complex With Moenomycin
Organism: Staphylococcus aureus mw2
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-06-27 Classification: TRANSFERASE Ligands: M0E, EDO, PO4, 1QW |
 |
Structure Of The Dna-Binding Domain Of Escherichia Coli Methionine Biosynthesis Regulator Metr
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2016-06-08 Classification: TRANSCRIPTION Ligands: MG, EDO |
 |
Crystal Structure Of Escherichia Coli 23S Rrna (A2030-N6)- Methyltransferase Rlmj
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2013-08-21 Classification: TRANSFERASE Ligands: TRS, GOL, EDO, SO4, PEG |
 |
Crystal Structure Of Escherichia Coli 23S Rrna (A2030-N6)- Methyltransferase Rlmj In Complex With S-Adenosylmethionine (Adomet)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-08-21 Classification: TRANSFERASE Ligands: SAM, GOL, PEG, EDO, SO4 |
 |
Crystal Structure Of Escherichia Coli 23S Rrna (A2030-N6)- Methyltransferase Rlmj In Complex With S-Adenosylhomocysteine (Adohcy) And Adenosine Monophosphate (Amp)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2013-08-21 Classification: TRANSFERASE Ligands: AMP, SAH, GOL, EDO, NA, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2012-12-05 Classification: STRUCTURAL PROTEIN Ligands: TRS, GOL |
 |
Crystal Structure Of C2498 2'-O-Ribose Methyltransferase Rlmm From Escherichia Coli
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2012-08-15 Classification: TRANSFERASE Ligands: GOL, EDO, SO4 |
 |
Crystal Structure Of C2498 2'-O-Ribose Methyltransferase Rlmm From Escherichia Coli
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2012-08-15 Classification: TRANSFERASE Ligands: GOL, SO4, TRS, TLA, PEG, PGE, EDO |
 |
Crystal Structure Of C2498 2'-O-Ribose Methyltransferase Rlmm From Escherichia Coli In Complex With S-Adenosylmethionine
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2012-08-15 Classification: TRANSFERASE Ligands: SAM, GOL, EDO, SO4 |

