Search Count: 16
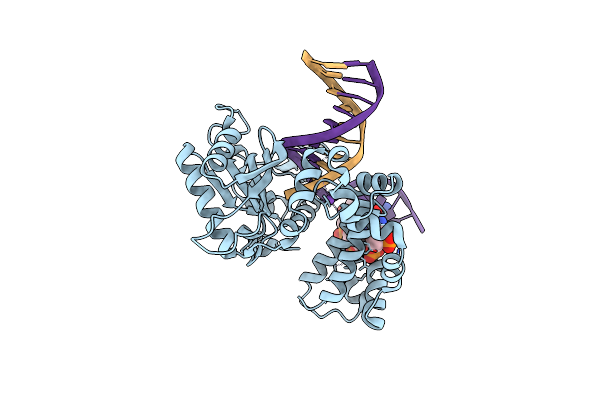 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-09-26 Classification: TRANSFERASE,LYASE/DNA |
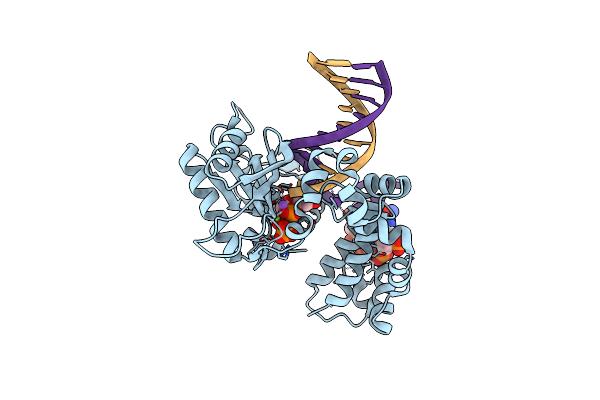 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2018-09-26 Classification: TRANSFERASE,LYASE/DNA |
 |
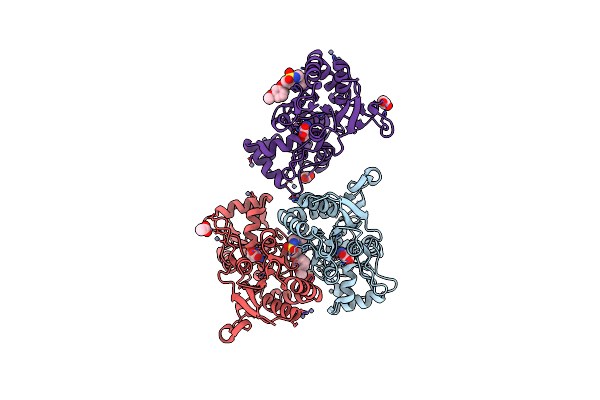
Crystal Structure Of The Glua2 Ligand-Binding Domain (S1S2J-L504Y-N775S) In Complex With Glutamate And Bpam121 At 1.78 A Resolution
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2018-01-03 Classification: MEMBRANE PROTEIN Ligands: SO4, 7M6, CL, GOL, GLU, EDO, CIT, PEG |
 |
Crystal Structure Of The Glua2 Ligand-Binding Domain (S1S2J) In Complex With Glutamate And Positive Allosteric Modulator Bpam538
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-01-03 Classification: MEMBRANE PROTEIN Ligands: GLU, ZN, ACT, 9TE |
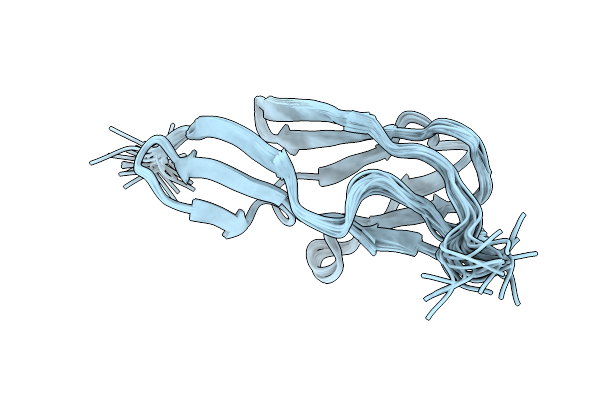 |
Solution Structure Of The Terminal Ig-Like Domain From Leptospira Interrogans Ligb
Organism: Leptospira interrogans
Method: SOLUTION NMR Release Date: 2014-08-13 Classification: CELL ADHESION |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2010-03-23 Classification: TRANSPORT PROTEIN Ligands: GLU, P99, ZN |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2010-03-23 Classification: TRANSPORT PROTEIN Ligands: GLU, ZN |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2010-03-23 Classification: TRANSPORT PROTEIN Ligands: GLU, P99, ZN |
 |
Crystal Structure Of The Ampa Subunit Glur2 Bound To The Allosteric Modulator, Althiazide
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-09-15 Classification: MEMBRANE PROTEIN Ligands: GLU, B4D, ZN |
 |
Crystal Structure Of The Ampa Subunit Glur2 Bound To The Allosteric Modulator, Hydrochlorothiazide
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.88 Å Release Date: 2009-09-15 Classification: MEMBRANE PROTEIN Ligands: GLU, HCZ, ZN |
 |
Crystal Structure Of The Ampa Subunit Glur2 Bound To The Allosteric Modulator, Chlorothiazide
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2009-09-15 Classification: MEMBRANE PROTEIN Ligands: GLU, HCZ, ZN |
 |
Crystal Structure Of The Ampa Subunit Glur2 Bound To The Allosteric Modulator, Idra-21
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-09-15 Classification: MEMBRANE PROTEIN Ligands: GLU, ZN, B5D |
 |
Crystal Structure Of The Ampa Subunit Glur2 Bound To The Allosteric Modulator, Trichlormethiazide
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2009-09-15 Classification: SIGNALING PROTEIN Ligands: GLU, ZN, TRU |
 |
Crystal Structure Of The Ampa Subunit Glur2 Bound To The Allosteric Modulator, Hydroflumethiazide
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-09-15 Classification: MEMBRANE PROTEIN Ligands: GLU, HFZ, ZN |
 |
Organism: Saccharomyces cerevisiae, Homo sapiens
Method: SOLUTION NMR Release Date: 2001-10-10 Classification: LIGASE |
 |
Crystal Structure Of Saccharomyces Cerevisiae Ubiquitin Conjugating Enzyme 1
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2001-10-04 Classification: LIGASE |

