Search Count: 21
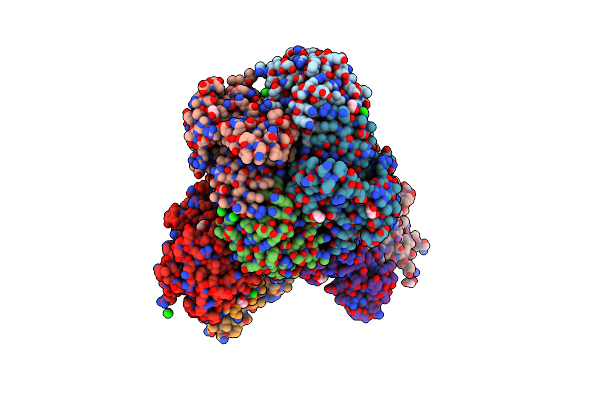 |
Crystal Structure Of Putataive Short-Chain Dehydrogenase/Reductase (Fabg) From Klebsiella Pneumoniae Subsp. Pneumoniae Ntuh-K2044 In Complex With Nadh
Organism: Klebsiella pneumoniae subsp. pneumoniae ntuh-k2044
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2022-03-02 Classification: BIOSYNTHETIC PROTEIN Ligands: K, CL, NAI, GOL, EDO |
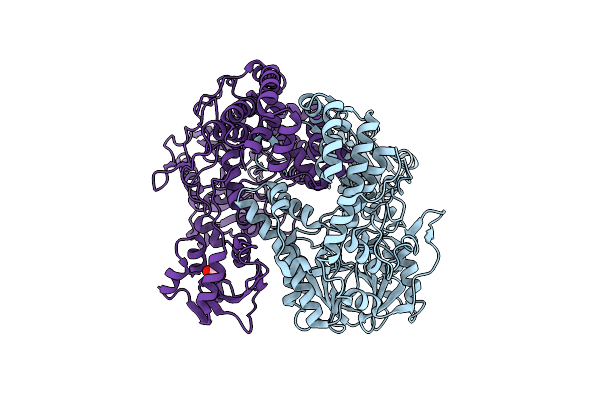 |
Organism: Klebsiella pneumoniae subsp. pneumoniae ntuh-k2044
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2022-02-02 Classification: HYDROLASE Ligands: EDO |
 |
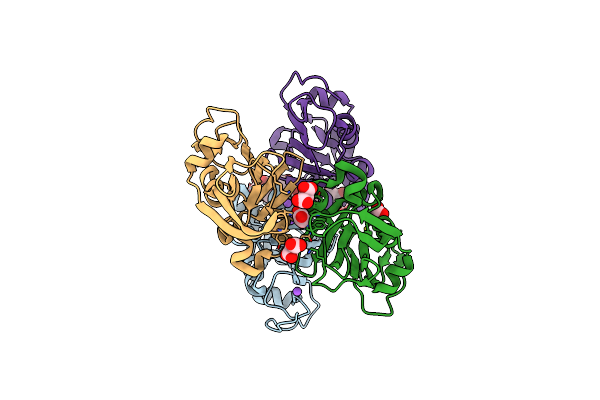
Crystal Structure Of The Succinyl-Diaminopimelate Desuccinylase (Dape) From Acinetobacter Baumannii In Complex With Succinic Acid
Organism: Acinetobacter baumannii atcc 17978
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2021-12-15 Classification: HYDROLASE Ligands: SIN, ACT, PGE, ZN |
 |
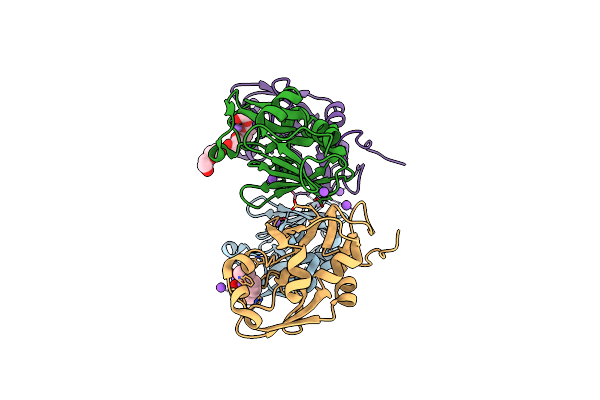
1.50 Angstroms Resolution Crystal Structure Of Putative Pterin Binding Protein Prur (Atu3496) From Agrobacterium Fabrum Str. C58
Organism: Agrobacterium fabrum (strain c58 / atcc 33970)
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2021-11-17 Classification: Pterin binding protein |
 |
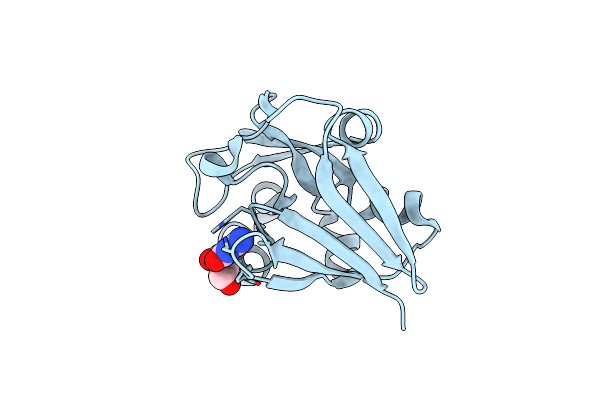
1.83 Angstroms Resolution Crystal Structure Of Putative Pterin Binding Protein Prur (Atu3496) From Agrobacterium Fabrum Str. C58
Organism: Agrobacterium fabrum (strain c58 / atcc 33970)
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2021-11-17 Classification: Pterin binding protein Ligands: CL, NA, PG4, PEG |
 |
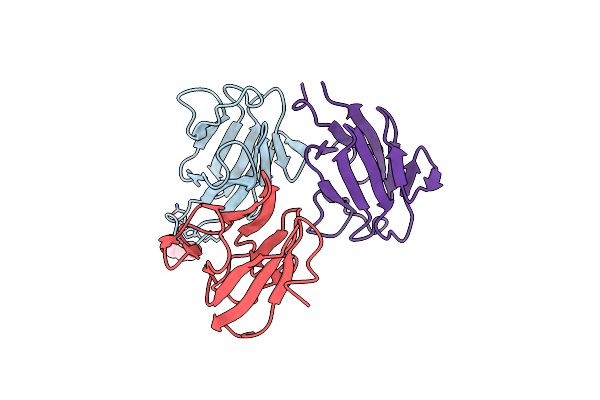
High Resolution Crystal Structure Of Putative Pterin Binding Protein (Prur) From Vibrio Cholerae O1 Biovar El Tor Str. N16961 In Complex With Neopterin
Organism: Vibrio cholerae o1 biovar el tor str. n16961
Method: X-RAY DIFFRACTION Resolution:1.03 Å Release Date: 2021-11-17 Classification: PTERIN-BINDING PROTEIN Ligands: NEU |
 |
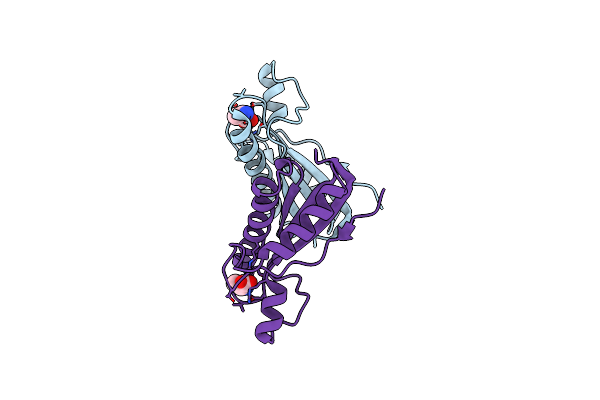
Crystal Structure Of The Forkhead Associated (Fha) Domain Of The Glycogen Accumulation Regulator (Gara) From Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2021-07-28 Classification: SIGNALING PROTEIN Ligands: PEG |
 |
Crystal Structure Of The Peptidoglycan Binding Domain Of The Outer Membrane Protein (Ompa) From Klebsiella Pneumoniae With Bound D-Alanine
Organism: Klebsiella pneumoniae subsp. pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2021-07-28 Classification: PEPTIDE BINDING PROTEIN Ligands: DAL, CL |
 |
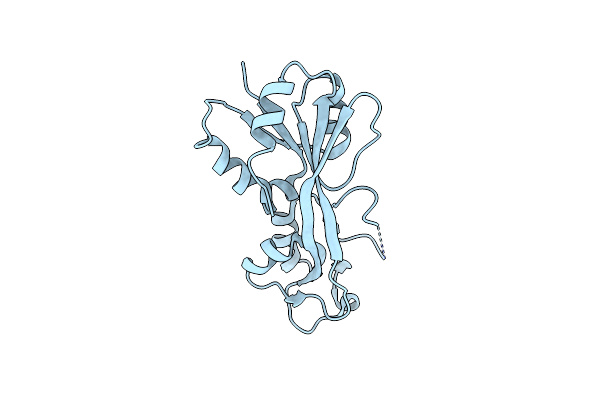
The Crystal Structure Of A Functional Uncharacterized Protein Kp1_0663 From Klebsiella Pneumoniae Subsp. Pneumoniae Ntuh-K2044
Organism: Klebsiella pneumoniae subsp. pneumoniae ntuh-k2044
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-06-03 Classification: UNKNOWN FUNCTION |
 |
1.52 Angstrom Resolution Crystal Structure Of Transcriptional Regulator Hdfr From Klebsiella Pneumoniae
Organism: Klebsiella pneumoniae subsp. pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2020-05-06 Classification: TRANSCRIPTION Ligands: CL |
 |
2.70 Angstrom Resolution Crystal Structure Of Uracil Phosphoribosyl Transferase From Klebsiella Pneumoniae
Organism: Klebsiella pneumoniae subsp. pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2020-05-06 Classification: TRANSFERASE Ligands: BDF, SO4, CL |
 |
Crystal Structure Of The Sugar Binding Domain Of Laci Family Protein From Klebsiella Pneumoniae
Organism: Klebsiella pneumoniae subsp. pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2018-12-26 Classification: SUGAR BINDING PROTEIN |
 |
1.55 Angstrom Resolution Crystal Structure Of 6-Phosphogluconolactonase From Klebsiella Pneumoniae
Organism: Klebsiella pneumoniae subsp. pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2018-12-19 Classification: HYDROLASE Ligands: B3P, CL |
 |
2.05 Angstrom Resolution Crystal Structure Of Hypothetical Protein Kp1_5497 From Klebsiella Pneumoniae.
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2018-12-19 Classification: UNKNOWN FUNCTION Ligands: PO4, CL |
 |
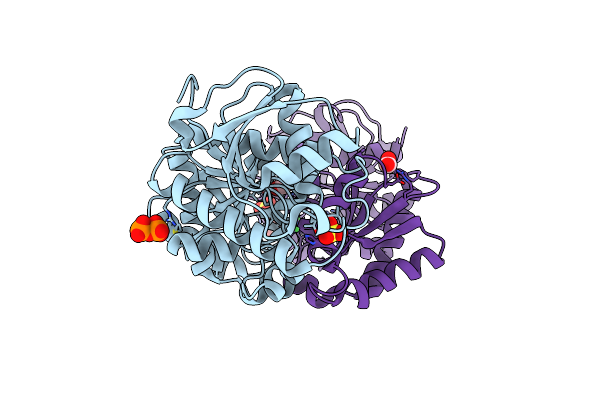
1.93 Angstrom Resolution Crystal Structure Of Peptidase M23 From Neisseria Gonorrhoeae.
Organism: Neisseria gonorrhoeae
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2018-10-31 Classification: HYDROLASE Ligands: ZN |
 |
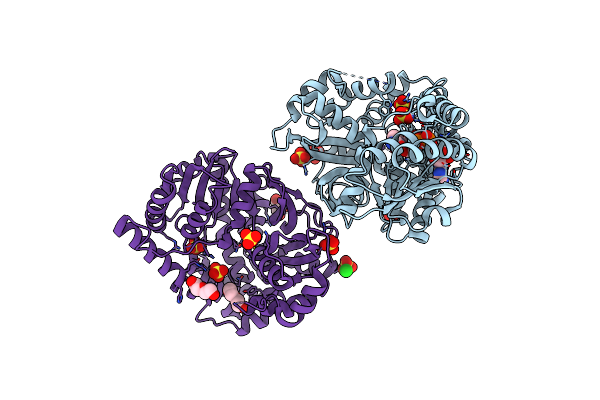
1.25 Angstrom Resolution Crystal Structure Of 4-Hydroxythreonine-4-Phosphate Dehydrogenase From Klebsiella Pneumoniae.
Organism: Klebsiella pneumoniae subsp. pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2018-08-08 Classification: HYDROLASE Ligands: NI, FMT, POP, SO4, CL |
 |
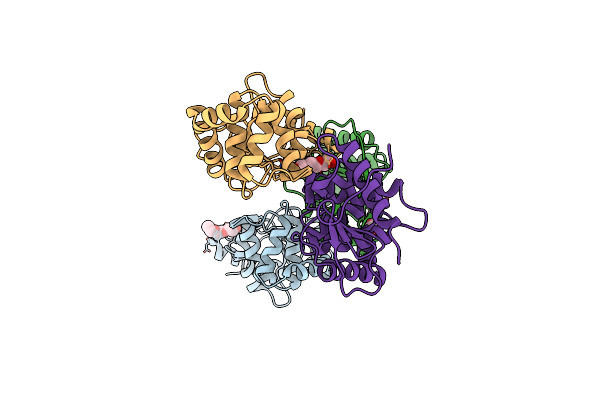
2.25 Angstrom Resolution Crystal Structure Of 6-Phospho-Alpha-Glucosidase From Klebsiella Pneumoniae In Complex With Nad And Mn2+.
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2018-07-18 Classification: HYDROLASE Ligands: NAD, MN, SO4, CL, PEG, GOL |
 |
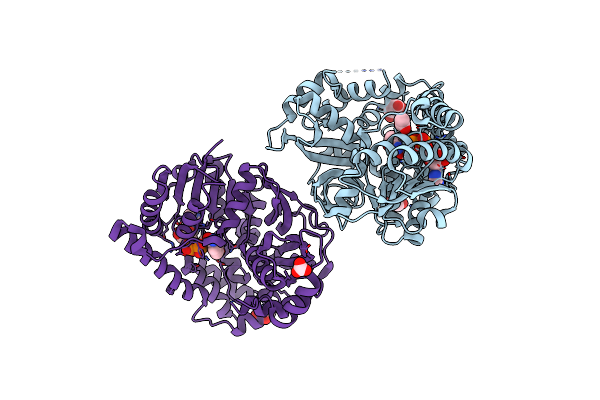
1.95 Angstrom Resolution Crystal Structure Of Dsba Disulfide Interchange Protein From Klebsiella Pneumoniae.
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2018-07-11 Classification: OXIDOREDUCTASE Ligands: PGE |
 |
2.25 Angstrom Resolution Crystal Structure Of 6-Phospho-Alpha-Glucosidase From Klebsiella Pneumoniae In Complex With Nad.
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2018-07-04 Classification: HYDROLASE Ligands: NAD, LMR, ACT, GOL, CL, BCT |
 |
1.2 Angstrom Resolution Crystal Structure Of Nucleoside Triphosphatase Nudi From Klebsiella Pneumoniae In Complex With Hepes
Organism: Klebsiella pneumoniae subsp. pneumoniae ntuh-k2044
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2018-06-27 Classification: HYDROLASE Ligands: EPE |

