Search Count: 13
 |
Crystal Structure Of P. Syringae Phosphinothricin Acetyltransferase Pspto_3321
Organism: Pseudomonas syringae pv. tomato str. dc3000
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-03-12 Classification: TRANSFERASE Ligands: CIT, PEG, NA, PGE |
 |
Crystal Structure Of P. Syringae Phosphinothricin Acetyltransferase Pspto_3321 In Complex With L-Phosphinothricin
Organism: Pseudomonas syringae pv. tomato str. dc3000
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2025-03-12 Classification: TRANSFERASE Ligands: SO4, PPQ, ACT, NA, EDO, PEG |
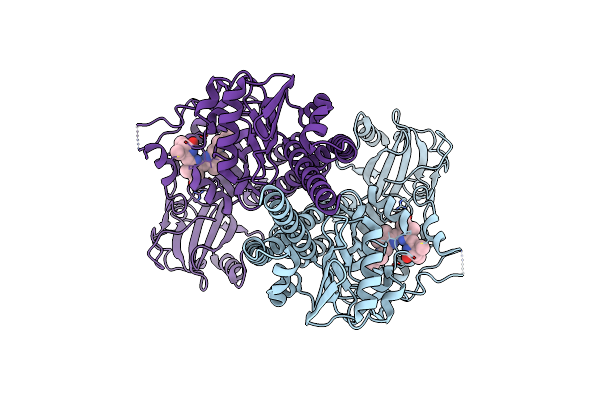 |
Organism: Pseudomonas syringae pv. tomato str. dc3000
Method: ELECTRON MICROSCOPY Release Date: 2024-08-21 Classification: PLANT PROTEIN Ligands: LBV |
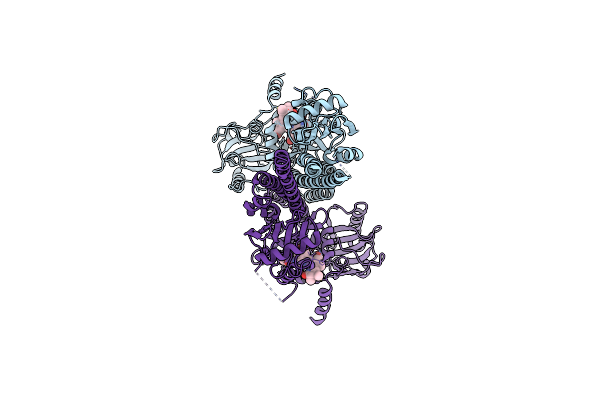 |
Organism: Pseudomonas syringae pv. tomato str. dc3000
Method: ELECTRON MICROSCOPY Release Date: 2024-08-21 Classification: PLANT PROTEIN Ligands: LBV |
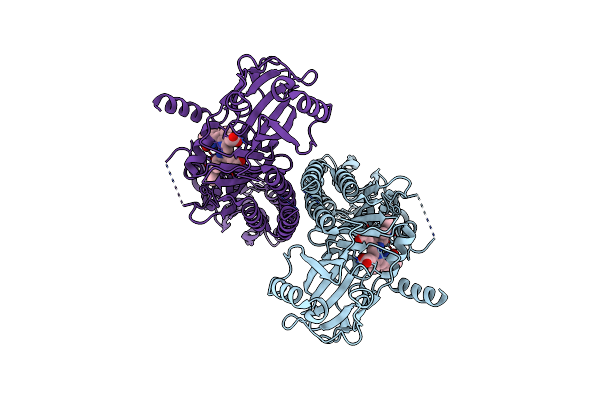 |
Organism: Pseudomonas syringae pv. tomato str. dc3000
Method: ELECTRON MICROSCOPY Release Date: 2024-08-21 Classification: PLANT PROTEIN Ligands: LBV |
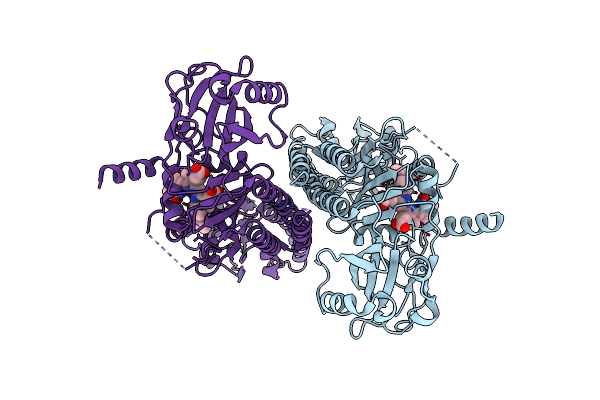 |
Organism: Pseudomonas syringae pv. tomato str. dc3000
Method: ELECTRON MICROSCOPY Release Date: 2024-08-21 Classification: PLANT PROTEIN Ligands: LBV |
 |
Organism: Pseudomonas syringae pv. tomato str. dc3000
Method: ELECTRON MICROSCOPY Release Date: 2024-08-21 Classification: PLANT PROTEIN Ligands: LBV |
 |
Organism: Pseudomonas syringae pv. tomato str. dc3000
Method: ELECTRON MICROSCOPY Release Date: 2024-08-21 Classification: PLANT PROTEIN Ligands: LBV |
 |
Organism: Pseudomonas syringae pv. tomato str. dc3000
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2022-10-05 Classification: LIGASE Ligands: ADP |
 |
Organism: Pseudomonas syringae pv. tomato str. dc3000
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2017-10-25 Classification: OXIDOREDUCTASE |
 |
The Structure Of The Effector Protein From Pseudomonas Syringae Pv. Tomato Strain Dc3000
Organism: Pseudomonas syringae pv. tomato str. dc3000
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2015-03-11 Classification: SIGNALING PROTEIN |
 |
Crystal Structure Of A Gtp-Regulated Atp Sulfurylase Heterodimer From Pseudomonas Syringae
Organism: Pseudomonas syringae, Pseudomonas syringae pv. tomato str. dc3000
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2006-01-17 Classification: TRANSFERASE Ligands: MG, NA, AGS, GDP |
 |
X-Ray Crystal Structure Of Protein Pspto5229 From Pseudomonas Syringae. Northeast Structural Genomics Consortium Target Psr62
Organism: Pseudomonas syringae pv. tomato str. dc3000
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2005-11-08 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION Ligands: MPO, EDO, 144 |

