Search Count: 769
 |
Crystal Structure Of P. Syringae Phosphinothricin Acetyltransferase Pspto_3321
Organism: Pseudomonas syringae pv. tomato str. dc3000
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-03-12 Classification: TRANSFERASE Ligands: CIT, PEG, NA, PGE |
 |
Crystal Structure Of P. Syringae Phosphinothricin Acetyltransferase Pspto_3321 In Complex With L-Phosphinothricin
Organism: Pseudomonas syringae pv. tomato str. dc3000
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2025-03-12 Classification: TRANSFERASE Ligands: SO4, PPQ, ACT, NA, EDO, PEG |
 |
Crystal Structure Of Endoribonuclease L-Psp Family Protein Pspto0102 Sulfur Sad Phased
Organism: Pseudomonas syringae pv. tomato
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2024-09-04 Classification: HYDROLASE |
 |
Organism: Pseudomonas syringae pv. tomato
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2024-09-04 Classification: HYDROLASE |
 |
Organism: Pseudomonas syringae pv. tomato str. dc3000
Method: ELECTRON MICROSCOPY Release Date: 2024-08-21 Classification: PLANT PROTEIN Ligands: LBV |
 |
Organism: Pseudomonas syringae pv. tomato str. dc3000
Method: ELECTRON MICROSCOPY Release Date: 2024-08-21 Classification: PLANT PROTEIN Ligands: LBV |
 |
Organism: Pseudomonas syringae pv. tomato str. dc3000
Method: ELECTRON MICROSCOPY Release Date: 2024-08-21 Classification: PLANT PROTEIN Ligands: LBV |
 |
Organism: Pseudomonas syringae pv. tomato str. dc3000
Method: ELECTRON MICROSCOPY Release Date: 2024-08-21 Classification: PLANT PROTEIN Ligands: LBV |
 |
Organism: Pseudomonas syringae pv. tomato str. dc3000
Method: ELECTRON MICROSCOPY Release Date: 2024-08-21 Classification: PLANT PROTEIN Ligands: LBV |
 |
Organism: Pseudomonas syringae pv. tomato str. dc3000
Method: ELECTRON MICROSCOPY Release Date: 2024-08-21 Classification: PLANT PROTEIN Ligands: LBV |
 |
Organism: Pseudomonas syringae pv. tomato str. dc3000
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2022-10-05 Classification: LIGASE Ligands: ADP |
 |
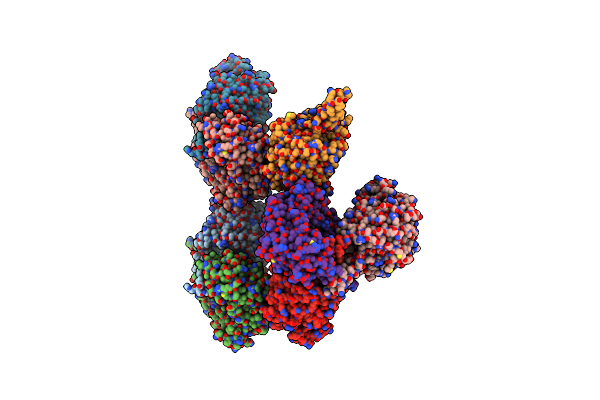
Structural Characterization Of L-Proline Amide Hydrolase From Pseudomonas Syringae
Organism: Pseudomonas syringae pv. tomato
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2021-10-06 Classification: HYDROLASE Ligands: PO4 |
 |
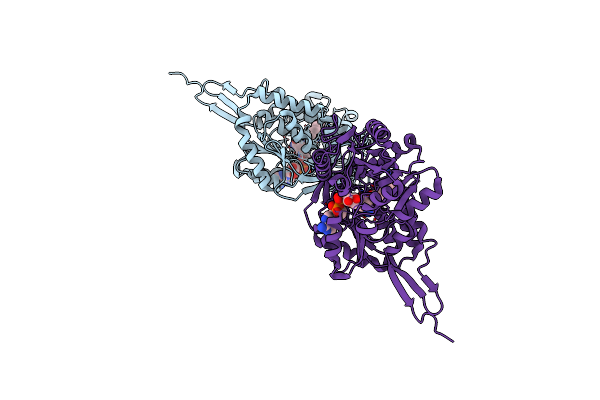
P. Syringae Alda Indole-3-Acetaldehyde Dehydrogenase C302A Mutant In Complex With Nad+ And Iaa
Organism: Pseudomonas syringae pv. tomato (strain atcc baa-871 / dc3000)
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2021-06-23 Classification: OXIDOREDUCTASE Ligands: NAI, IAC |
 |
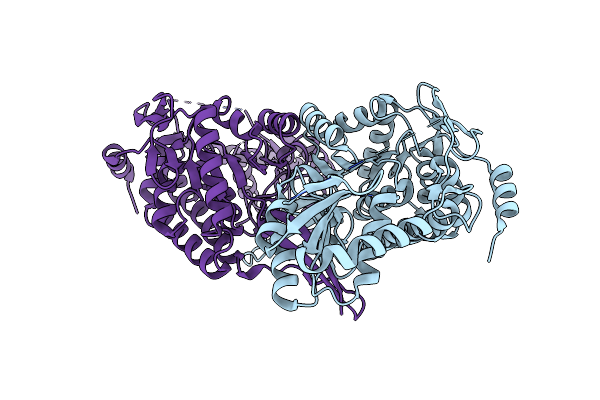
Crystal Structure Of Aldehyde Dehydrogenase C (Aldc) Mutant (C291A) From Pseudomonas Syringae In Complexed With Nad+ And Octanal
Organism: Pseudomonas syringae pv. tomato (strain atcc baa-871 / dc3000)
Method: X-RAY DIFFRACTION Resolution:2.52 Å Release Date: 2020-09-09 Classification: OXIDOREDUCTASE Ligands: OYA, NAD |
 |
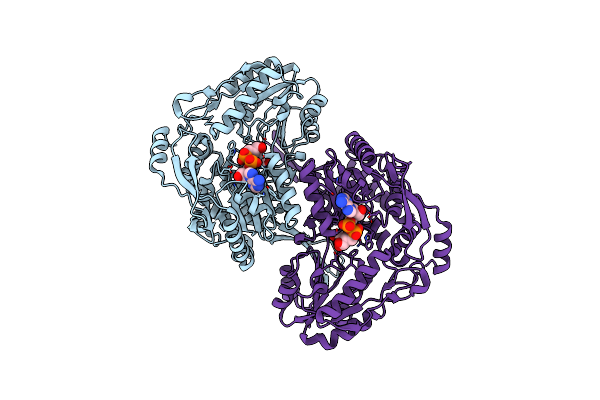
Organism: Pseudomonas syringae pv. tomato str. dc3000
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2017-10-25 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of Indole-3-Acetaldehyde Dehydrogenase In Complexed With Nad+
Organism: Pseudomonas syringae pv. tomato (strain dc3000)
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2017-10-25 Classification: OXIDOREDUCTASE Ligands: NAD |
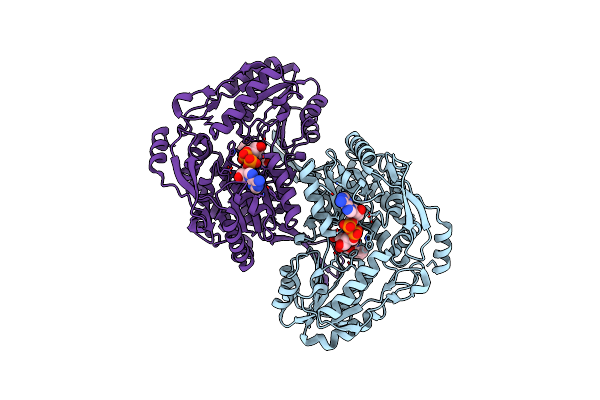 |
Crystal Structure Of Indole-3-Acetaldehyde Dehydrogenase In Complexed With Nad+ And Iaa
Organism: Pseudomonas syringae pv. tomato (strain dc3000)
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2017-10-25 Classification: OXIDOREDUCTASE Ligands: NAD, IAC |
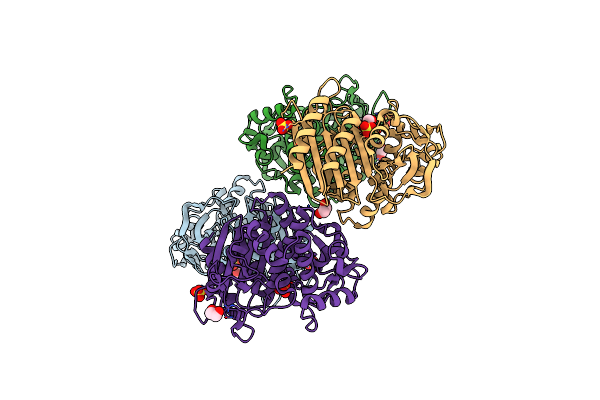 |
Crystal Structure Of Beta-Lactamase/D-Alanine Carboxypeptidase From Pseudomonas Syringae
Organism: Pseudomonas syringae pv. tomato
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-01-13 Classification: HYDROLASE Ligands: SO4, EDO |
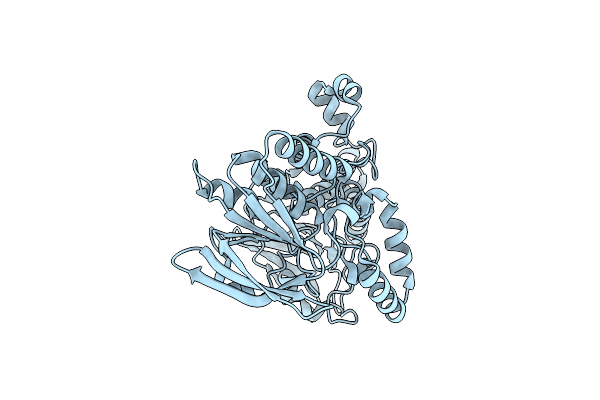 |
Organism: Pseudomonas syringae pv. tomato (strain dc3000)
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2015-09-30 Classification: OXIDOREDUCTASE Ligands: IOD |
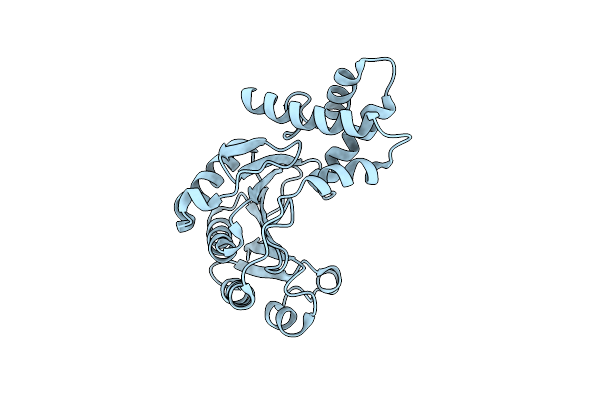 |
The Structure Of The Effector Protein From Pseudomonas Syringae Pv. Tomato Strain Dc3000
Organism: Pseudomonas syringae pv. tomato str. dc3000
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2015-03-11 Classification: SIGNALING PROTEIN |

