Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
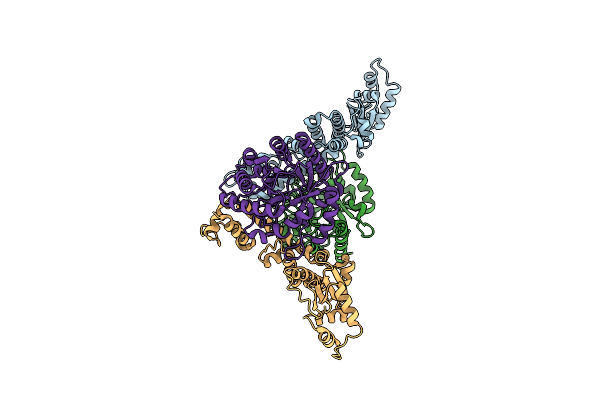 |
Crystal Structure Of A Holoenzyme Fe-Free Tglhi For Pseudomonas Syringae Peptidyl (S) 2-Mercaptoglycine Biosynthesis
Organism: Pseudomonas syringae pv. maculicola str. es4326
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2023-08-23 Classification: BIOSYNTHETIC PROTEIN |
 |
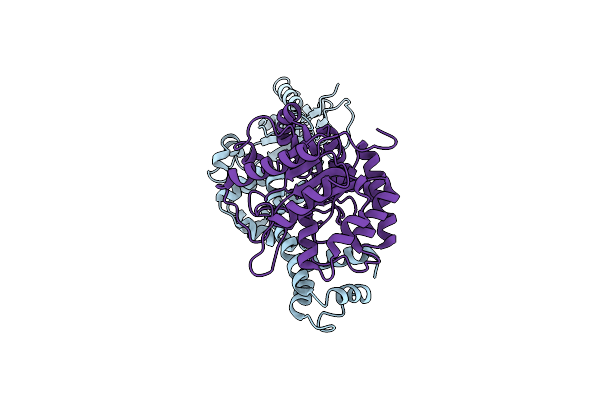
Crystal Structure Of A Holoenzyme Tglhi With Two Fe Irons For Pseudomonas Syringae Peptidyl (S) 2-Mercaptoglycine Biosynthesis
Organism: Pseudomonas syringae pv. maculicola str. es4326
Method: X-RAY DIFFRACTION Resolution:3.25 Å Release Date: 2023-08-23 Classification: METAL BINDING PROTEIN Ligands: FE |
 |
Crystal Structure Of A Holoenzyme Tglhi With Three Fe Ions For Pseudomonas Syringae Peptidyl (S) 2-Mercaptoglycine Biosynthesis
Organism: Pseudomonas syringae pv. maculicola str. es4326
Method: X-RAY DIFFRACTION Resolution:3.49 Å Release Date: 2023-08-23 Classification: METAL BINDING PROTEIN Ligands: FE |
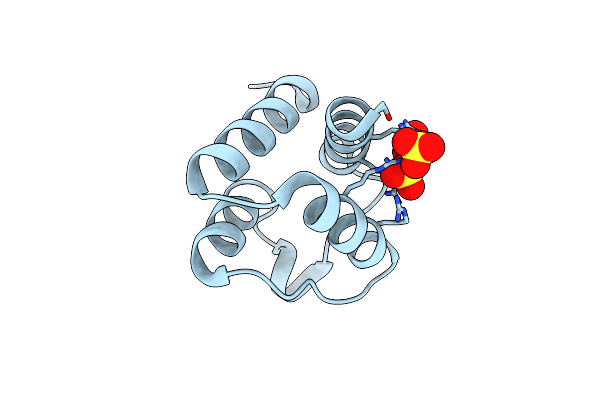 |
Structure Of The Pto-Binding Domain Of Hoppmal Generated By Limited Chymotrypsin Digestion
Organism: Pseudomonas syringae pv. maculicola
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2011-09-14 Classification: SIGNALING PROTEIN Ligands: SO4, CL |
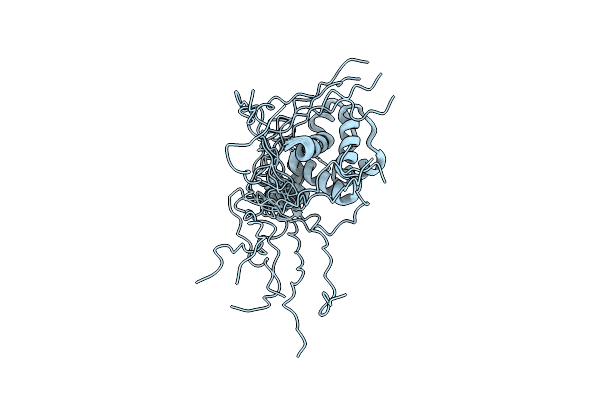 |
Solution Nmr Structure Of Hoppmal_281_385 From Pseudomonas Syringae Pv. Maculicola Str. Es4326, Midwest Center For Structural Genomics Target Apc40104.5 And Northeast Structural Genomics Consortium Target Pst2A
Organism: Pseudomonas syringae pv. maculicola
Method: SOLUTION NMR Release Date: 2011-07-13 Classification: SIGNALING PROTEIN |
 |
NA
Organism: Pseudomonas syringae pv. maculicola
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
NA
Organism: Pseudomonas syringae pv. maculicola
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
NA
Organism: Pseudomonas syringae pv. maculicola
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
NA
Organism: Pseudomonas syringae pv. maculicola
Method: Alphafold Release Date: Classification: NA Ligands: NA |