Search Count: 1,994
 |
Organism: Pseudomonas sp. mc1
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: UNKNOWN FUNCTION |
 |
Organism: Pseudomonas sp. uk4
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: PROTEIN FIBRIL |
 |
Crystal Structure Of Methyl Parathion Hydrolase Mutant A127L/I145L/Q272D/S279A/S304P
Organism: Pseudomonas sp. wbc-3
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: HYDROLASE Ligands: ZN |
 |
Crystal Structure Of Methyl Parathion Hydrolase Mutant A66D/I143V/I145L/Q272D/S279A/
Organism: Pseudomonas sp. wbc-3
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: HYDROLASE Ligands: ZN |
 |
Crystal Structure Of The Enzyme-Product Complex Of L-Azetidine-2-Carboxylate Hydrolase
Organism: Pseudomonas sp. a2c
Method: X-RAY DIFFRACTION Resolution:0.93 Å Release Date: 2025-06-04 Classification: HYDROLASE Ligands: 42B, IMD, FMT, EDO, PEG, MG |
 |
Organism: Pseudomonas sp.
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-12-11 Classification: FLAVOPROTEIN Ligands: ALA, FAD, GLY |
 |
Crystal Structure Of Acetylpolyamine Amidohydrolase (Apah) From Pseudomonas Sp. M30-35
Organism: Pseudomonas sp. m30-35
Method: X-RAY DIFFRACTION Resolution:2.72 Å Release Date: 2024-11-06 Classification: HYDROLASE Ligands: PGE, K, ZN, CL, ACT, PEG |
 |
Organism: Pseudomonas sp. mis38
Method: SOLUTION NMR Release Date: 2024-08-14 Classification: SURFACTANT PROTEIN |
 |
Organism: Pseudomonas sp.
Method: ELECTRON MICROSCOPY Release Date: 2024-06-26 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Organism: Pseudomonas sp.
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2024-06-26 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Organism: Pseudomonas sp. a2c
Method: X-RAY DIFFRACTION Resolution:1.19 Å Release Date: 2024-05-08 Classification: HYDROLASE Ligands: FMT, IMD, MG |
 |
Crystal Structure Of L-Azetidine-2-Carboxylate Hydrolase Soaked In (S)-Azetidine-2-Carboxylic Acid
Organism: Pseudomonas sp. a2c
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2024-05-08 Classification: HYDROLASE Ligands: 02A |
 |
Cryo-Em Structure Of Styrene Oxide Isomerase Bound To Benzylamine Inhibitor
Organism: Pseudomonas sp. vlb120, Vicugna pacos
Method: ELECTRON MICROSCOPY Release Date: 2024-04-03 Classification: MEMBRANE PROTEIN Ligands: ABN, HEM |
 |
Organism: Pseudomonas sp. vlb120, Vicugna pacos
Method: ELECTRON MICROSCOPY Release Date: 2024-04-03 Classification: MEMBRANE PROTEIN Ligands: HEM |
 |
Organism: Pseudomonas sp. 'mevalonii'
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2024-03-20 Classification: STRUCTURAL PROTEIN Ligands: COA, MEV, SO4, NAD |
 |
Organism: Pseudomonas sp. 'mevalonii'
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-03-20 Classification: OXIDOREDUCTASE Ligands: ZKE, SO4, XII |
 |
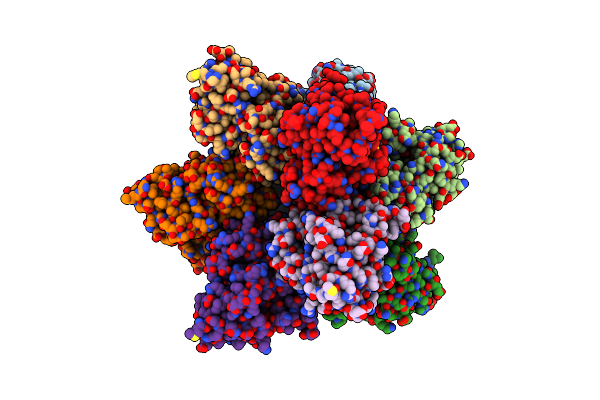
Structure Of Pmhmgr Bound To Mevalonate, Coa And Nad 5 Minutes After Reaction Initiation At Ph 9
Organism: Pseudomonas sp. 'mevalonii'
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2024-03-20 Classification: OXIDOREDUCTASE Ligands: ZKE, COA, A1AFY, SO4, NAD, NAI, MEV |
 |
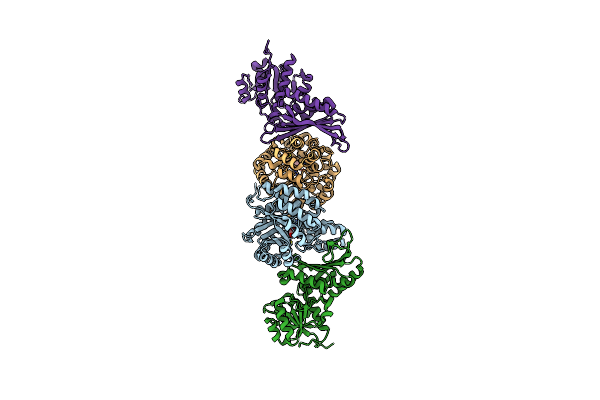
Crystal Structure Of Decarboxylase-Hydratase Complex From Pseudomonas Species Ap-3
Organism: Pseudomonas sp.
Method: X-RAY DIFFRACTION Resolution:2.52 Å Release Date: 2024-02-28 Classification: BIOSYNTHETIC PROTEIN Ligands: GOL |
 |
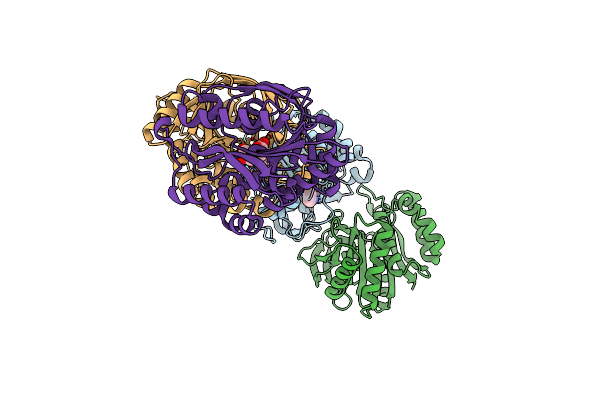
Organism: Pseudomonas sp.
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2024-02-28 Classification: BIOSYNTHETIC PROTEIN Ligands: ZN, OXL |
 |
Organism: Pseudomonas sp.
Method: X-RAY DIFFRACTION Resolution:2.86 Å Release Date: 2024-02-28 Classification: BIOSYNTHETIC PROTEIN Ligands: ZN, 2AF, GOL |

