Search Count: 34
 |
Organism: Providencia stuartii
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2024-12-25 Classification: RNA |
 |
Structure Of E. Coli Glpg In Complex With Peptide Derived Inhibitor Ac-Vrha-Conh-[4-(4-Butyl)-Phenoxy-1-Phenyl-2-Butyl]
Organism: Escherichia coli k-12, Providencia stuartii
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2023-10-25 Classification: HYDROLASE |
 |
Organism: Providencia stuartii
Method: ELECTRON MICROSCOPY Release Date: 2022-04-20 Classification: LYASE |
 |
Organism: Providencia stuartii
Method: ELECTRON MICROSCOPY Release Date: 2022-04-20 Classification: LYASE |
 |
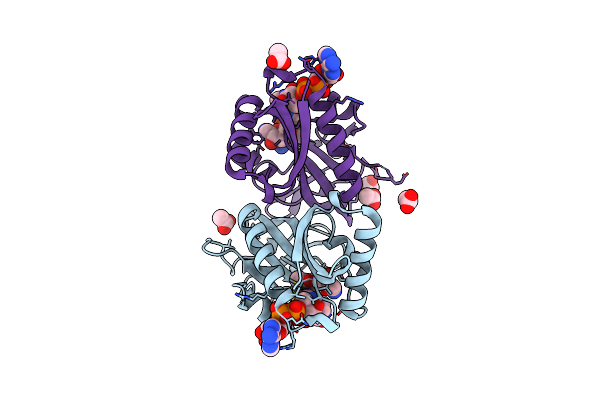
Aminoglycoside N-2'-Acetyltransferase-Ia [Aac(2')-Ia] In Complex With Acetylated-Plazomicin And Coa
Organism: Providencia stuartii
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2021-06-02 Classification: TRANSFERASE Ligands: COA, TCE, MPD, PZC |
 |
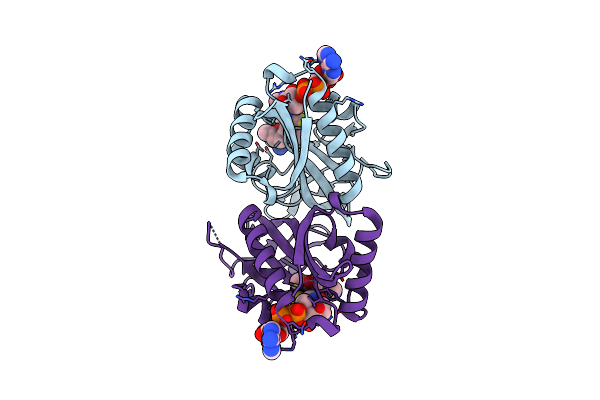
Aminoglycoside N-2'-Acetyltransferase-Ia [Aac(2')-Ia] In Complex With Acetylated-Tobramycin And Coa
Organism: Providencia stuartii
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2021-06-02 Classification: TRANSFERASE Ligands: R7Y, COA, ACT |
 |
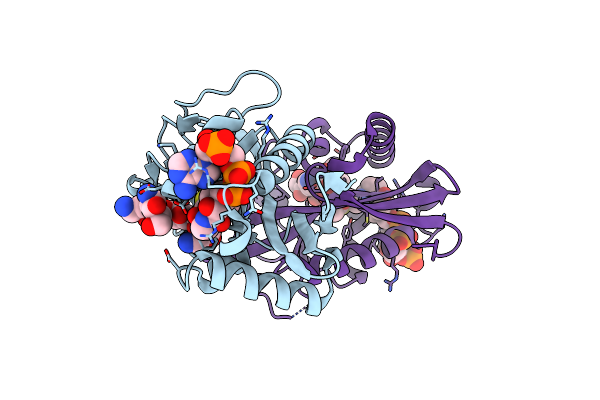
Aminoglycoside N-2'-Acetyltransferase-Ia [Aac(2')-Ia] In Complex With Acetylated-Netilmicin And Coa
Organism: Providencia stuartii
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-06-02 Classification: TRANSFERASE Ligands: NTL, COA |
 |
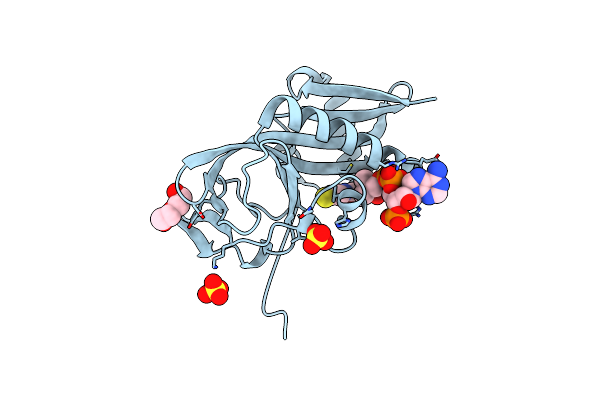
Aminoglycoside N-2'-Acetyltransferase-Ia [Aac(2')-Ia] In Complex With Amikacin And Acetyl-Coa
Organism: Providencia stuartii
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2021-06-02 Classification: TRANSFERASE Ligands: ACO, AKN |
 |
Organism: Providencia stuartii
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2021-06-02 Classification: TRANSFERASE Ligands: COA, SO4, PEG |
 |
Structure Of 283-Lgny-286, The Steric Zipper That Supports The Self-Association Of P. Stuartii Omp-Pst2 Into Dimers Of Trimers
Organism: Providencia stuartii
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2018-02-21 Classification: CELL ADHESION Ligands: SO4 |
 |
Structure Of 206-Gvvtse-211, The Steric Zipper That Supports The Self-Association Of P. Stuartii Omp-Pst1 Into Dimers Of Trimers
Organism: Providencia stuartii
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2018-02-21 Classification: CELL ADHESION |
 |
Organism: Providencia stuartii
Method: X-RAY DIFFRACTION Resolution:3.12 Å Release Date: 2018-02-21 Classification: TRANSPORT PROTEIN Ligands: CA |
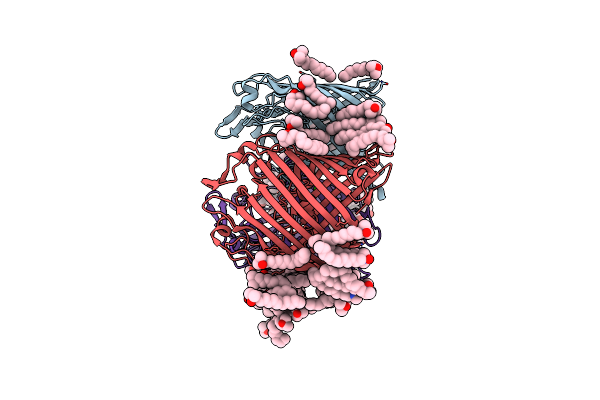 |
Organism: Providencia stuartii
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2018-02-21 Classification: CELL ADHESION Ligands: LDA, CA, CL |
 |
Organism: Providencia stuartii
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2018-02-21 Classification: CELL ADHESION Ligands: LDA, CA |
 |
Structure Of E.Coli Glpg In Complex With Peptide Derived Inhibitor Ac-Rvrha-Phenylethyl-Ketoamide
Organism: Escherichia coli k-12, Providencia stuartii
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2017-11-15 Classification: HYDROLASE Ligands: BNG |
 |
Structure Of E.Coli Glpg In Complex With Peptide Derived Inhibitor Ac-Vrha-Cmk
Organism: Escherichia coli k-12, Providencia stuartii
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2017-11-15 Classification: HYDROLASE |
 |
Structure Of E.Coli Glpg In Complex With Peptide Derived Inhibitor Ac-Rvrha-Cmk
Organism: Escherichia coli k-12, Providencia stuartii
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2017-11-15 Classification: HYDROLASE Ligands: BNG, CL |
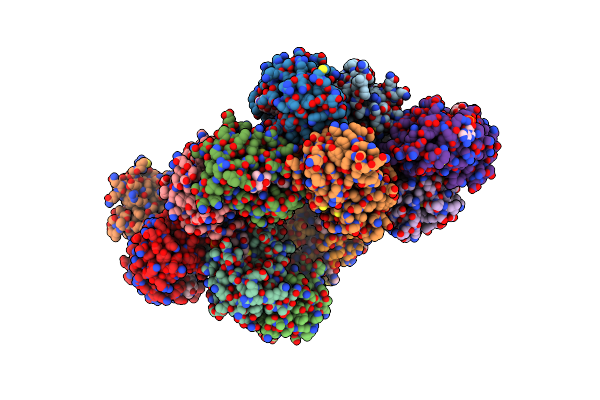 |
Crystal Structure Of Aminoglycoside Acetyltransferase Aac(2')-Ia In Complex With N2'-Acetylgentamicin C1A And Coenzyme A
Organism: Providencia stuartii
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2017-03-15 Classification: TRANSFERASE Ligands: 8MM, TAR, ACO, GOL, COA |
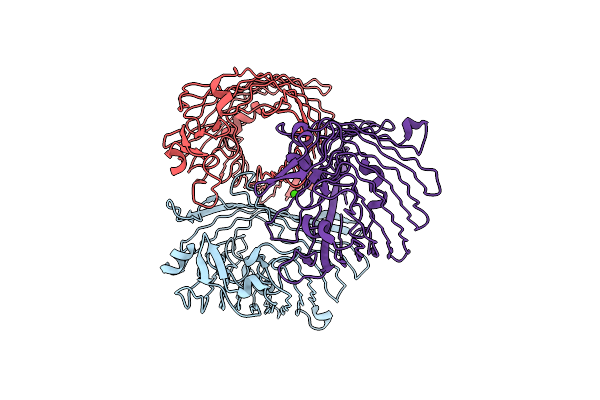 |
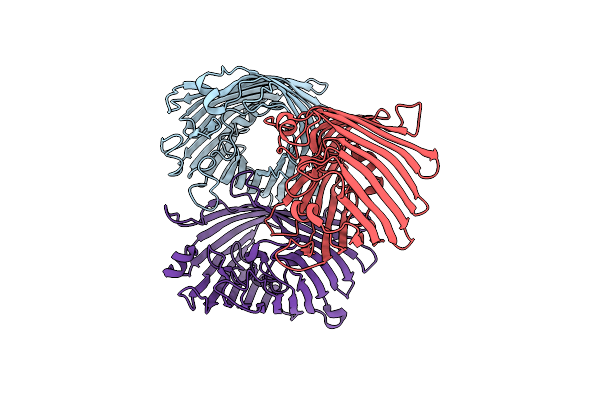
Structure Of Porin Omp-Pst1 From P. Stuartii; The Crystallographic Symmetry Generates A Dimer Of Trimers.
Organism: Providencia stuartii
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2016-03-09 Classification: TRANSPORT PROTEIN Ligands: CA |
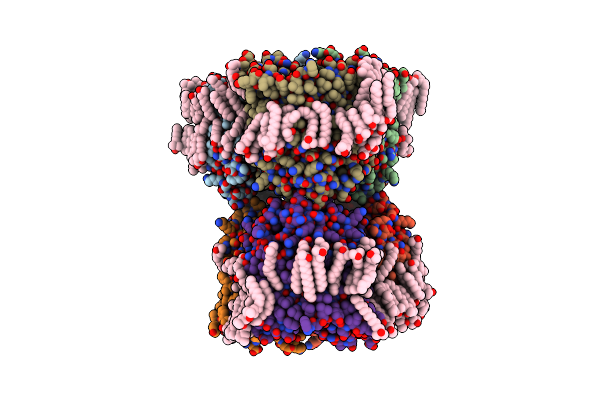 |
Structure Of Porin Omp-Pst2 From P. Stuartii; The Asymmetric Unit Contains A Dimer Of Trimers.
Organism: Providencia stuartii
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2016-03-09 Classification: TRANSPORT PROTEIN Ligands: LDA, FTT, MYR, SO4 |

