Search Count: 1,452
All
Selected
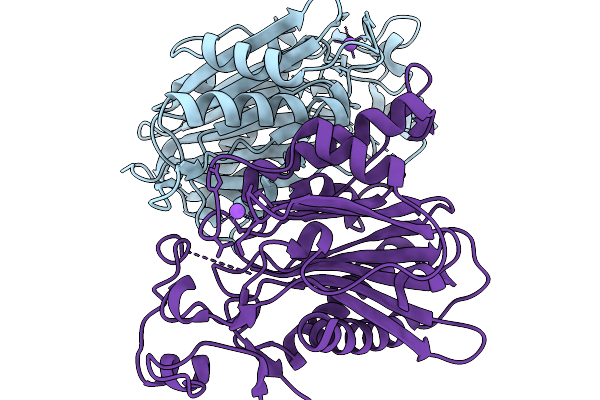 |
Crystal Structure Of The Poly(Hexamethylene Adipamide) (Nylon66) Hydrolase Nyl50 In Its Apo Form At Room Temperature
Organism: Alphaproteobacteria bacterium
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-11-19 Classification: HYDROLASE |
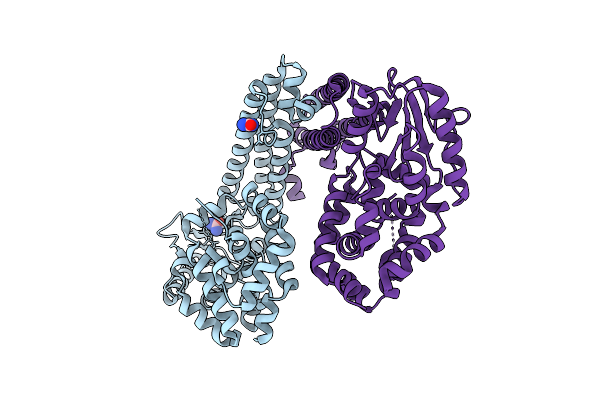 |
Organism: Gammaproteobacteria
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2025-08-06 Classification: DNA BINDING PROTEIN Ligands: URE |
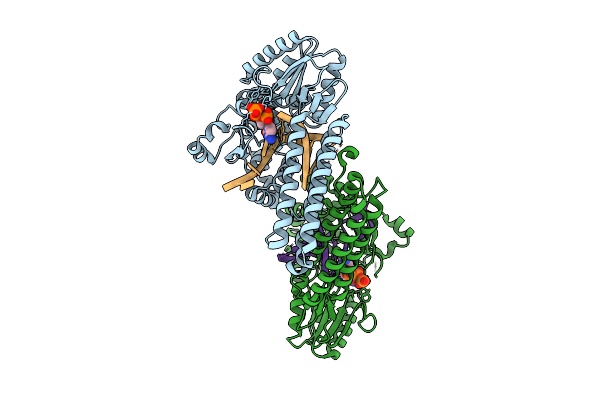 |
Organism: Gammaproteobacteria, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.77 Å Release Date: 2025-07-30 Classification: DNA BINDING PROTEIN Ligands: MN, DCP |
 |
X-Ray Crystallographic Structure Of The Poly(Hexamethylene Adipamide) (Nylon66) Hydrolase Nyl50 At Room Temperature
Organism: Alphaproteobacteria bacterium
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-01-29 Classification: HYDROLASE Ligands: EDO, NA, PGE |
 |
X-Ray Crystallographic Structure Of The Poly(Hexamethylene Adipamide) (Nylon66) Hydrolase Nyl50 At Room Temperature Bound To Tetraethylene Glycol
Organism: Alphaproteobacteria bacterium
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2025-01-29 Classification: HYDROLASE Ligands: EDO, NA, PG4 |
 |
Organism: Deltaproteobacteria
Method: ELECTRON MICROSCOPY Release Date: 2025-01-15 Classification: IMMUNE SYSTEM/RNA |
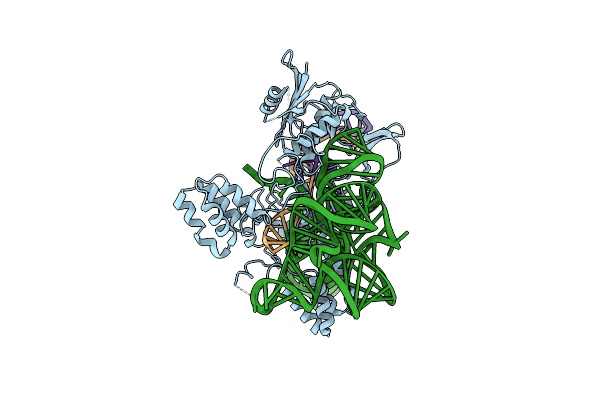 |
Organism: Deltaproteobacteria, Escherichia phage lambda
Method: ELECTRON MICROSCOPY Release Date: 2025-01-15 Classification: IMMUNE SYSTEM/RNA/DNA |
 |
Organism: Deltaproteobacteria
Method: ELECTRON MICROSCOPY Release Date: 2025-01-15 Classification: DNA BINDING PROTEIN, RNA BINDING PROTEIN/DNA/RNA |
 |
Crystal Structure Of 2-Hydroxacyl-Coa Lyase/Synthase Apbhacs From Alphaproteobacteria Bacterium In The Complex With Thdp, L-Lactyl-Coa, And Adp
Organism: Alphaproteobacteria bacterium
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2024-10-02 Classification: LYASE Ligands: 8FL, UQ3, ADP, ACE, MG, GOL |
 |
Crystal Structure Of 2-Hydroxacyl-Coa Lyase/Synthase Apbhacs From Alphaproteobacteria Bacterium In The Complex With Thdp, D-Lactyl-Coa, And Adp
Organism: Alphaproteobacteria bacterium
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2024-10-02 Classification: LYASE Ligands: MG, 8FL, UQ3, ADP, EDO, ACY, PO4, CL, ACE, PEG |
 |
Crystal Structure Of 2-Hydroxyacyl-Coa Lyase/Synthse Apbhacs From Alphaproteobacteria Bacterium In The Complex With Thdp, Formyl-Coa, And Adp
Organism: Alphaproteobacteria bacterium
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2024-10-02 Classification: LYASE Ligands: A1AEK, FYN, MG, ADP, EDO, PO4, PEG |
 |
Crystal Structure Of 2-Hydroxyacyl-Coa Lyase/Synthase Apbhacs From Alphaproteobacteria Bacterium In The Complex With Thdp, Coenzyme A, And Adp
Organism: Alphaproteobacteria bacterium
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2024-10-02 Classification: LYASE Ligands: FYN, TPP, MG, ADP, EDO, SO4, CL, GOL |
 |
Organism: Uncultured gammaproteobacteria bacterium
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: PROTON TRANSPORT Ligands: DKA |
 |
Organism: Uncultured gammaproteobacteria bacterium
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: PROTON TRANSPORT Ligands: RET |
 |
Organism: Uncultured gammaproteobacteria bacterium
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: PROTON TRANSPORT Ligands: RET |
 |
Organism: Gammaproteobacteria bacterium
Method: X-RAY DIFFRACTION Resolution:2.63 Å Release Date: 2021-11-17 Classification: UNKNOWN FUNCTION Ligands: BTN, GOL |
 |
Organism: Gammaproteobacteria bacterium
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2021-11-17 Classification: UNKNOWN FUNCTION |
 |
Organism: Gammaproteobacteria bacterium
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2021-11-17 Classification: UNKNOWN FUNCTION Ligands: 1PE, P6G, PEG, PGE |
 |
Organism: Gammaproteobacteria bacterium
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2021-11-17 Classification: UNKNOWN FUNCTION Ligands: BTN, PG4, PGE, EDO, PEG, P33, NA |
 |
Organism: Epsilonproteobacteria bacterium (ex lamellibrachia satsuma)
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2021-10-27 Classification: HYDROLASE Ligands: MN |

