Search Count: 3,957
All
Selected
 |
Organism: Prochlorococcus marinus subsp. pastoris str. ccmp1986
Method: NEUTRON DIFFRACTION Resolution:2.10 Å Release Date: 2024-01-17 Classification: METAL BINDING PROTEIN Ligands: FE |
 |
Crystal Structure Of Urta From Prochlorococcus Marinus Str. Mit 9313 In Complex With Urea And Calcium
Organism: Prochlorococcus marinus str. mit 9313
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-11-22 Classification: TRANSPORT PROTEIN Ligands: URE, CA |
 |
Organism: Prochlorococcus marinus subsp. pastoris str. ccmp1986
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-08-30 Classification: METAL BINDING PROTEIN Ligands: FE |
 |
Organism: Prochlorococcus marinus subsp. pastoris str. ccmp1986
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-08-30 Classification: METAL BINDING PROTEIN Ligands: FE |
 |
Organism: Prochlorococcus marinus subsp. pastoris str. ccmp1986
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-08-30 Classification: METAL BINDING PROTEIN Ligands: FE2 |
 |
Organism: Prochlorococcus marinus subsp. pastoris str. ccmp1986
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2023-08-30 Classification: METAL BINDING PROTEIN Ligands: FE |
 |
Solution Nmr Structure Of Prochlorosin 2.11 (Pcn2.11) Produced By Prochlorococcus Mit 9313
Organism: Prochlorococcus marinus str. mit 9313
Method: SOLUTION NMR Release Date: 2020-09-09 Classification: UNKNOWN FUNCTION |
 |
Solution Nmr Structure Of Prochlorosin 2.10 Produced By Prochlorococcus Mit 9313
Organism: Prochlorococcus marinus str. mit 9313
Method: SOLUTION NMR Release Date: 2020-09-09 Classification: UNKNOWN FUNCTION |
 |
Organism: Prochlorococcus marinus str. mit 9313
Method: SOLUTION NMR Release Date: 2020-07-08 Classification: UNKNOWN FUNCTION |
 |
Solution Nmr Structure Of Prochlorosin 2.1 Produced By Prochlorococcus Mit 9313
Organism: Prochlorococcus marinus str. mit 9313
Method: SOLUTION NMR Release Date: 2020-07-08 Classification: UNKNOWN FUNCTION |
 |
Organism: Prochlorococcus marinus str. mit 9313
Method: SOLUTION NMR Release Date: 2020-07-08 Classification: UNKNOWN FUNCTION |
 |
Organism: Prochlorococcus marinus (strain mit 9303)
Method: SOLUTION NMR Release Date: 2019-08-21 Classification: RNA BINDING PROTEIN |
 |
Organism: Prochlorococcus marinus str. mit 9302, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2019-07-10 Classification: DNA BINDING PROTEIN Ligands: MN, AMP, SO4 |
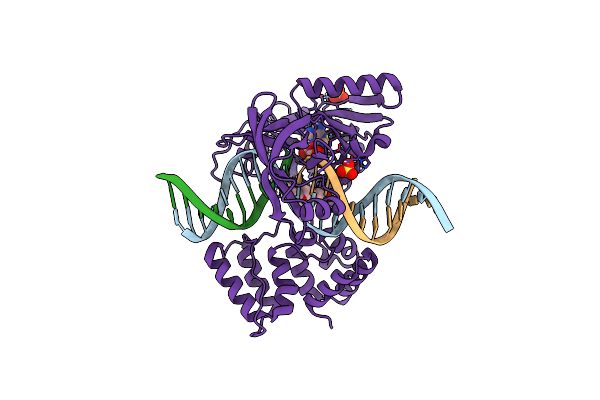 |
Organism: Prochlorococcus marinus str. mit 9302, Dna launch vector pde-gfp2
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2019-07-10 Classification: DNA BINDING PROTEIN Ligands: PGE, AMP, SO4 |
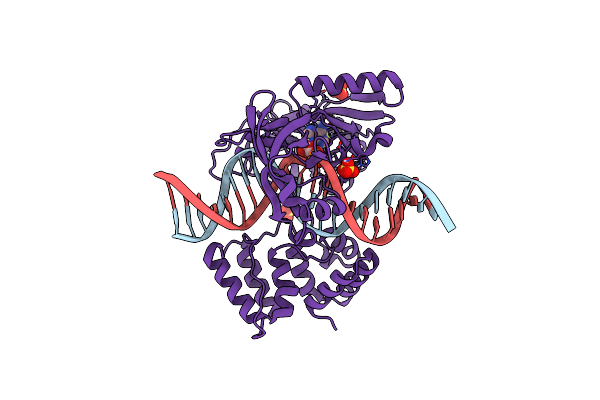 |
Organism: Prochlorococcus marinus, Dna launch vector pde-gfp2
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2019-07-10 Classification: DNA BINDING PROTEIN Ligands: AMP, SO4 |
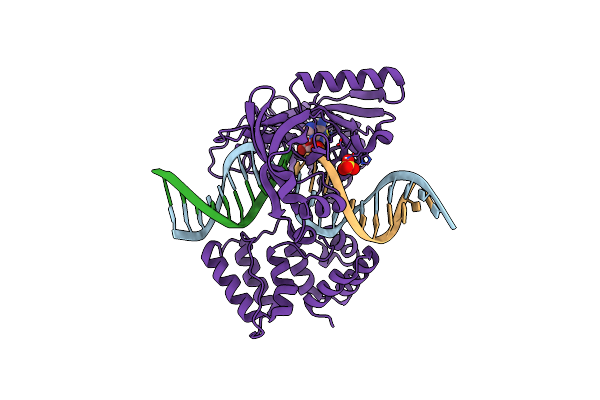 |
Organism: Prochlorococcus marinus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2019-07-10 Classification: DNA BINDING PROTEIN Ligands: AMP, SO4 |
 |
Crystal Structure Of A Double Glycine Motif Protease From Ams/Pcat Transporter In Complex With The Leader Peptide
Organism: Lachnospiraceae bacterium c6a11, Prochlorococcus marinus str. mit 9313
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-02-06 Classification: TRANSPORT PROTEIN Ligands: 16P |
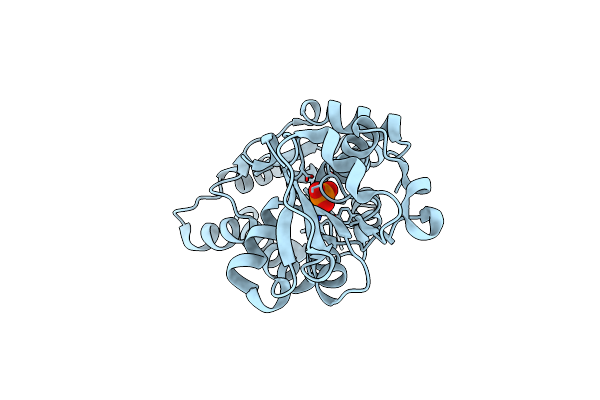 |
1.46 A Resolution Structure Of Phnd1 From Prochlorococcus Marinus (Mit 9301) In Complex With Phosphite
Organism: Prochlorococcus marinus str. mit 9301
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2017-12-06 Classification: TRANSPORT PROTEIN Ligands: PO3 |
 |
1.52 A Resolution Structure Of Phnd1 From Prochlorococcus Marinus (Mit 9301) In Complex With Methylphosphonate
Organism: Prochlorococcus marinus str. mit 9301
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2017-12-06 Classification: PERIPLASMIC BINDING PROTEIN Ligands: GB |
 |
2.12 A Resolution Structure Of Ptxb From Prochlorococcus Marinus (Mit 9301) In Complex With Phosphite
Organism: Prochlorococcus marinus (strain mit 9301)
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2017-12-06 Classification: PERIPLASMIC BINDING PROTEIN Ligands: 78T |

