Search Count: 33
 |
Organism: Homo sapiens, Vicugna pacos huacaya
Method: ELECTRON MICROSCOPY Release Date: 2023-10-11 Classification: MEMBRANE PROTEIN Ligands: NAG |
 |
Human Alpha7 Nicotinic Receptor In Complex With The C4 Nanobody And Nicotine
Organism: Homo sapiens, Vicugna pacos
Method: ELECTRON MICROSCOPY Release Date: 2023-10-11 Classification: MEMBRANE PROTEIN Ligands: NAG, NCT |
 |
Organism: Homo sapiens, Vicugna pacos
Method: ELECTRON MICROSCOPY Release Date: 2023-10-11 Classification: MEMBRANE PROTEIN Ligands: NAG |
 |
Human Alpha7 Nicotinic Receptor In Complex With The E3 Nanobody And Nicotine
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-10-11 Classification: MEMBRANE PROTEIN Ligands: NAG, NCT |
 |
Human Alpha7 Nicotinic Receptor In Complex With The C4 Nanobody Under Sub-Saturating Conditions
Organism: Homo sapiens, Vicugna pacos
Method: ELECTRON MICROSCOPY Release Date: 2023-10-11 Classification: MEMBRANE PROTEIN Ligands: NAG |
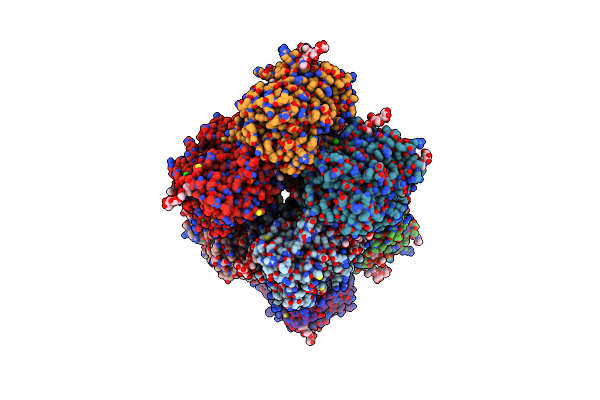 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2022-08-24 Classification: ANTIMICROBIAL PROTEIN Ligands: CL, CA, HEM, SCN, 8PR, PO4, NAG |
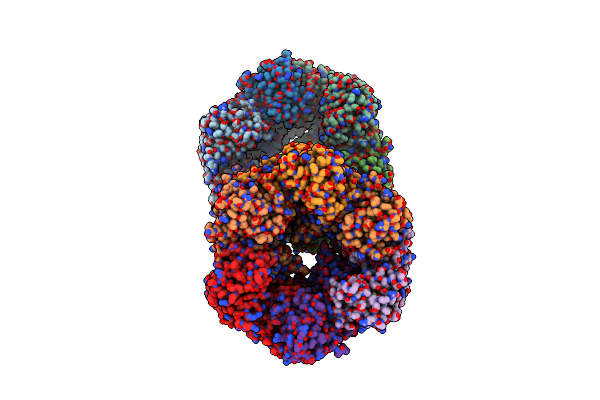 |
Organism: Legionella pneumophila
Method: X-RAY DIFFRACTION Resolution:3.19 Å Release Date: 2018-05-30 Classification: HYDROLASE Ligands: PO4 |
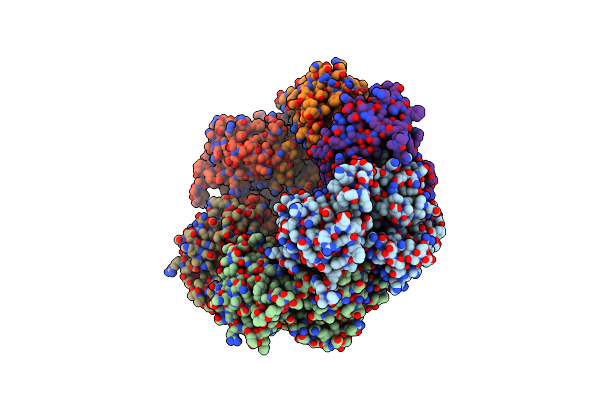 |
Organism: Yersinia pseudotuberculosis ip 31758
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2018-05-30 Classification: HYDROLASE |
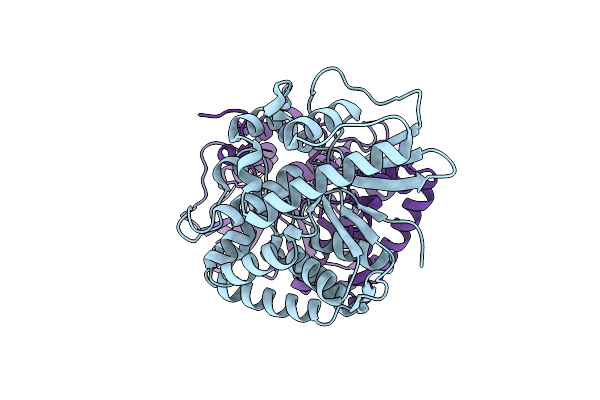 |
Crystal Structure Of The Phosphatase Domain From The Legionella Effector Wipb
Organism: Legionella pneumophila
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2017-09-06 Classification: HYDROLASE |
 |
Breaking Down The Wall: Mutation Of The Tyrosine Gate Of The Universal Escherichia Coli Fimbrial Adhesin Fimh
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-01-18 Classification: CELL ADHESION Ligands: EDT |
 |
Breaking Down The Wall: Mutation Of The Tyrosine Gate Of The Universal Escherichia Coli Fimbrial Adhesin Fimh
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2016-11-30 Classification: CELL ADHESION Ligands: KGM, NA |
 |
Breaking Down The Wall: Mutation Of The Tyrosine Gate Of The Universal Escherichia Coli Fimbrial Adhesin Fimh
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2016-11-30 Classification: CELL ADHESION Ligands: 3X8 |
 |
Nmr Structure Of A 180 Residue Construct Encompassing The N-Terminal Metal-Binding Site And The Membrane Proximal Domain Of Silb From Cupriavidus Metallidurans Ch34
Organism: Cupriavidus metallidurans
Method: SOLUTION NMR Release Date: 2016-05-18 Classification: METAL BINDING PROTEIN Ligands: AG |
 |
Crystal Structure Of Fimh Lectin Domain With The Tyr48Ala Mutation, In Complex With Heptyl Alpha-D-Mannopyrannoside
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.84 Å Release Date: 2014-10-29 Classification: CELL ADHESION Ligands: KGM |
 |
The Glic Pentameric Ligand-Gated Ion Channel E19'P Mutant In A Locally-Closed Conformation (Lc2 Subtype)
Organism: Gloeobacter violaceus
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2012-05-16 Classification: MEMBRANE PROTEIN, TRANSPORT PROTEIN Ligands: CL, LMT |
 |
The Glic Pentameric Ligand-Gated Ion Channel H11'F Mutant In A Locally-Closed Conformation (Lc1 Subtype)
Organism: Gloeobacter violaceus
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2012-05-16 Classification: MEMBRANE PROTEIN, TRANSPORT PROTEIN Ligands: CL, LMT |
 |
The Glic Pentameric Ligand-Gated Ion Channel Loop2-24' Oxidized Mutant In A Locally-Closed Conformation (Lc1 Subtype)
Organism: Gloeobacter violaceus
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2012-05-16 Classification: MEMBRANE PROTEIN, TRANSPORT PROTEIN Ligands: CL, LMT |
 |
The Glic Pentameric Ligand-Gated Ion Channel Loop2-22' Oxidized Mutant In A Locally-Closed Conformation (Lc3 Subtype)
Organism: Gloeobacter violaceus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2012-05-16 Classification: MEMBRANE PROTEIN, TRANSPORT PROTEIN Ligands: LMT, CL |
 |
The Glic Pentameric Ligand-Gated Ion Channel Loop2-21' Oxidized Mutant In A Locally-Closed Conformation (Lc2 Subtype)
Organism: Gloeobacter violaceus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2012-05-16 Classification: MEMBRANE PROTEIN, TRANSPORT PROTEIN Ligands: CL, LMT |
 |
The Glic Pentameric Ligand-Gated Ion Channel Loop2-20' Oxidized Mutant In A Locally-Closed Conformation (Lc1 Subtype)
Organism: Gloeobacter violaceus
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2012-05-16 Classification: MEMBRANE PROTEIN, TRANSPORT PROTEIN Ligands: LMT |

