Search Count: 22
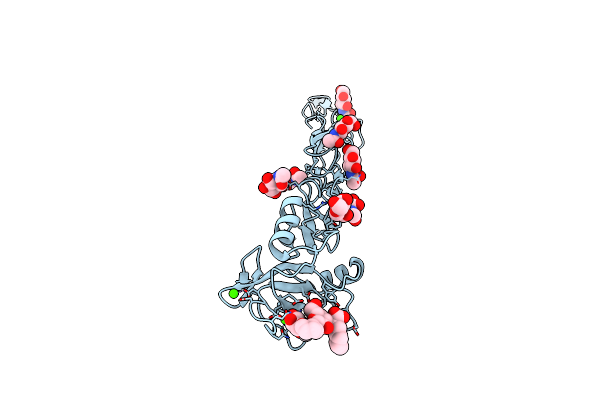 |
E-Selectin Lectin, Egf-Like And Two Scr Domains Complexed With Glycomimetic Ligand Bw69669
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2018-11-21 Classification: CELL ADHESION Ligands: NAG, C4Z, CA |
 |
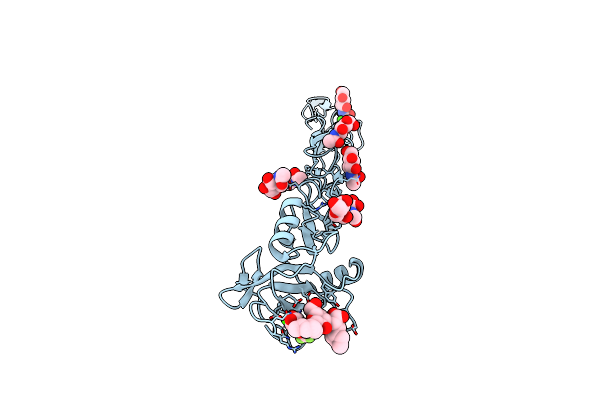
E-Selectin Lectin, Egf-Like And Two Scr Domains Complexed With Glycomimetic Ligand Nv354
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-11-21 Classification: CELL ADHESION Ligands: NAG, C5H, CA |
 |
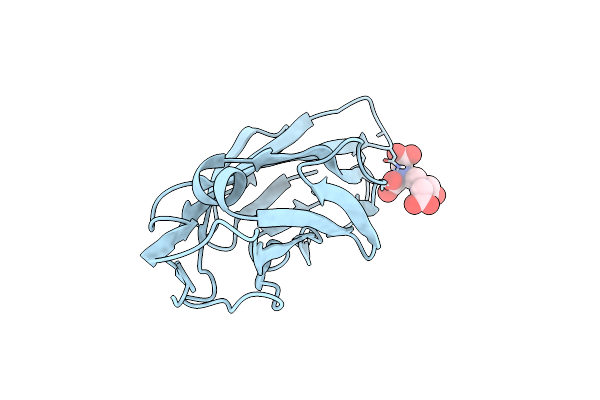
E-Selectin Lectin, Egf-Like And Two Scr Domains Complexed With Glycomimetic Ligand Nv355
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2018-11-21 Classification: CELL ADHESION Ligands: NAG, C5K, CA |
 |
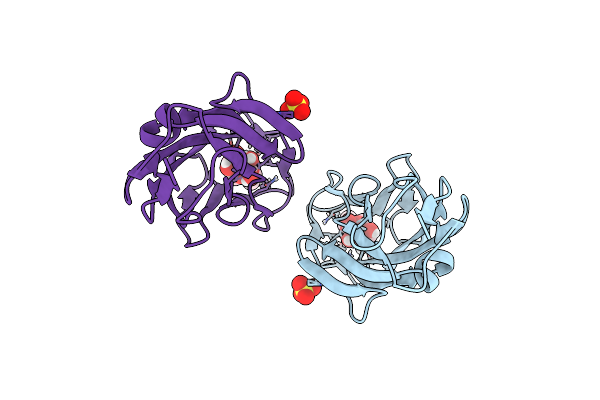
Breaking Down The Wall: Mutation Of The Tyrosine Gate Of The Universal Escherichia Coli Fimbrial Adhesin Fimh
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-01-18 Classification: CELL ADHESION Ligands: EDT |
 |
Breaking Down The Wall: Mutation Of The Tyrosine Gate Of The Universal Escherichia Coli Fimbrial Adhesin Fimh
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2016-11-30 Classification: CELL ADHESION Ligands: KGM, NA |
 |
Breaking Down The Wall: Mutation Of The Tyrosine Gate Of The Universal Escherichia Coli Fimbrial Adhesin Fimh
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2016-11-30 Classification: CELL ADHESION Ligands: 3X8 |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2016-07-20 Classification: SUGAR BINDING PROTEIN Ligands: 51C, SO4 |
 |
Crystal Structure Of The Lectin Domain Of Papg From E. Coli Bi47 In Complex With Ssea4 In Space Group P212121
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-04-13 Classification: SUGAR BINDING PROTEIN |
 |
Crystal Structure Of The Lectin Domain Of Papg From E. Coli Bi47 In Complex With Ssea4 In Space Group P21
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-04-13 Classification: SUGAR BINDING PROTEIN Ligands: ZN |
 |
Crystal Structure Of The Lectin Domain Of Papg From E. Coli Bi47 In Complex With 4-Methoxyphenyl Beta-D-Galabiose In Space Group P212121
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2016-04-13 Classification: SUGAR BINDING PROTEIN Ligands: MES, 4KS |
 |
Crystal Structure Of The Lectin Domain Of Papg From E. Coli Bi47 In Complex With 4-Methoxyphenyl Beta-D-Galabiose In Space Group P21
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2016-04-13 Classification: SUGAR BINDING PROTEIN Ligands: 4KS |
 |
Crystal Structure Of The Lectin Domain Of Papg From E. Coli Bi47 In Spacegroup P21212
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2016-04-13 Classification: SUGAR BINDING PROTEIN |
 |
Crystal Structure Of The Lectin Domain Of Papg From E. Coli Bi47 In Space Group P1
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2016-04-13 Classification: SUGAR BINDING PROTEIN |
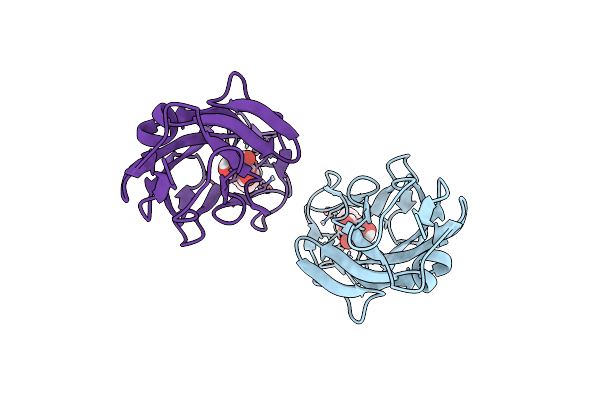 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-05-20 Classification: SUGAR BINDING PROTEIN Ligands: 3X8 |
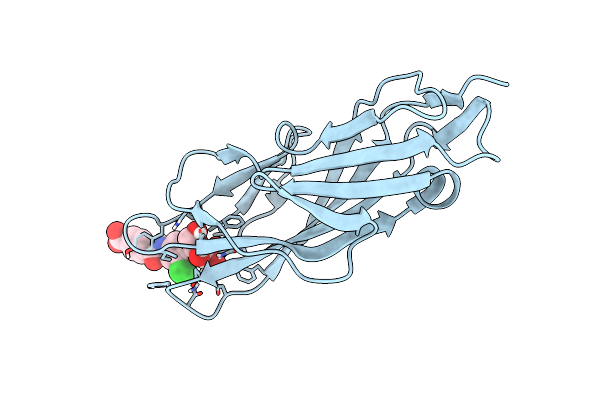 |
Crystal Structure Of Fimh In Complex With A Benzoyl-Amidophenyl Alpha-D-Mannopyranoside
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2015-05-20 Classification: SUGAR BINDING PROTEIN Ligands: 3XJ |
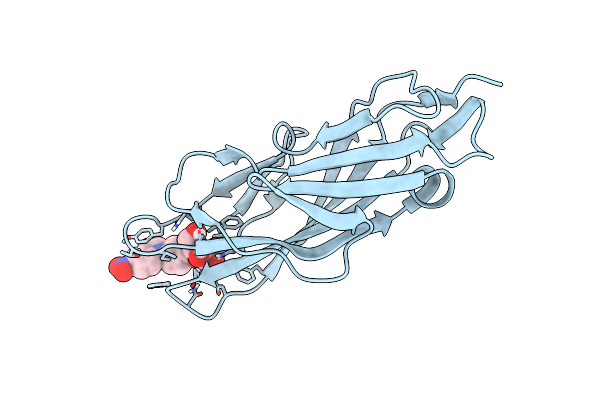 |
Crystal Structure Of Fimh In Complex With 5-Nitro-Indolinylphenyl Alpha-D-Mannopyranoside
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.12 Å Release Date: 2015-05-20 Classification: SUGAR BINDING PROTEIN Ligands: 3XN |
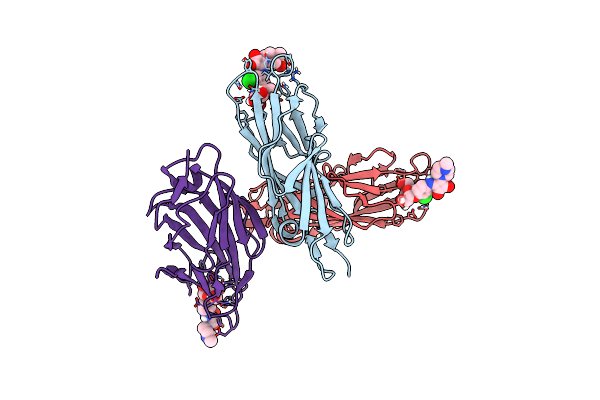 |
Crystal Structure Of Fimh In Complex With A Squaryl-Phenyl Alpha-D-Mannopyranoside Derivative
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2015-05-20 Classification: SUGAR BINDING PROTEIN Ligands: 3XO, SO4 |
 |
Crystal Structure Of Fimh In Complex With A Sulfonamide Biphenyl Alpha D-Mannoside
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.07 Å Release Date: 2015-02-25 Classification: SUGAR BINDING PROTEIN Ligands: CWX, GOL |
 |
Crystal Structure Of Fimh In Complex With 3'-Chloro-4'-(Alpha-D-Mannopyranosyloxy)-Biphenyl-4-Carbonitrile
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2015-02-25 Classification: SUGAR BINDING PROTEIN Ligands: CWK |
 |
Crystal Structure Of Fimh Lectin Domain With The Tyr48Ala Mutation, In Complex With Heptyl Alpha-D-Mannopyrannoside
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.84 Å Release Date: 2014-10-29 Classification: CELL ADHESION Ligands: KGM |

