Search Count: 7
 |
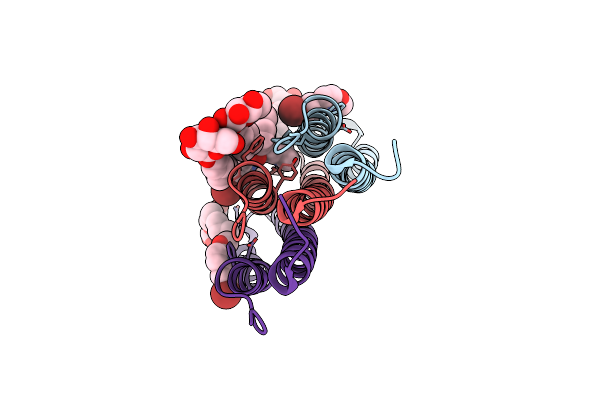
Crystal Structure Of A Mycobacterial Atp Synthase Rotor Ring In Complex With Iodo-Bedaquiline
Organism: Mycobacterium phlei
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-01-20 Classification: HYDROLASE Ligands: IBQ, TAM |
 |
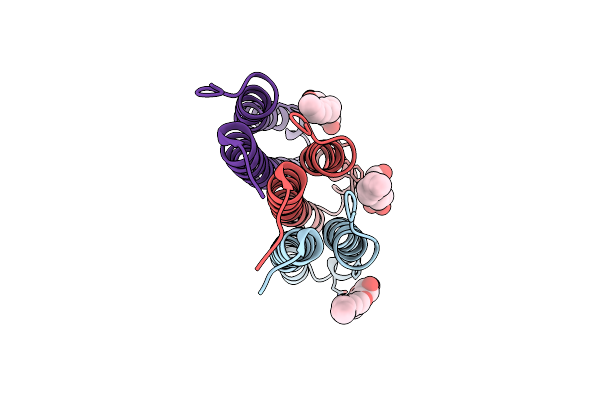
Crystal Structure Of A Mycobacterial Atp Synthase Rotor Ring In Complex With Bedaquiline
Organism: Mycobacterium phlei
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2015-05-20 Classification: HYDROLASE Ligands: BQ1, BOG |
 |
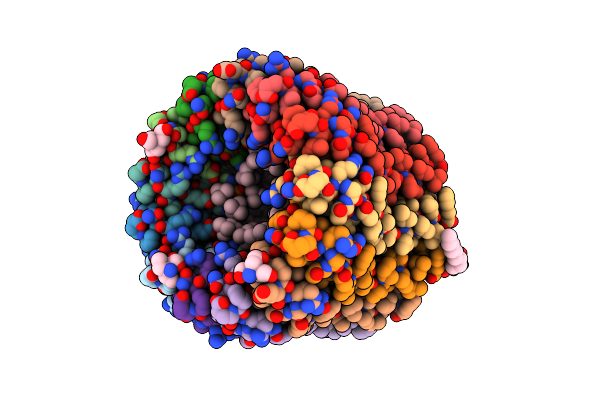
Organism: Mycobacterium phlei
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2015-05-20 Classification: HYDROLASE Ligands: BOG |
 |
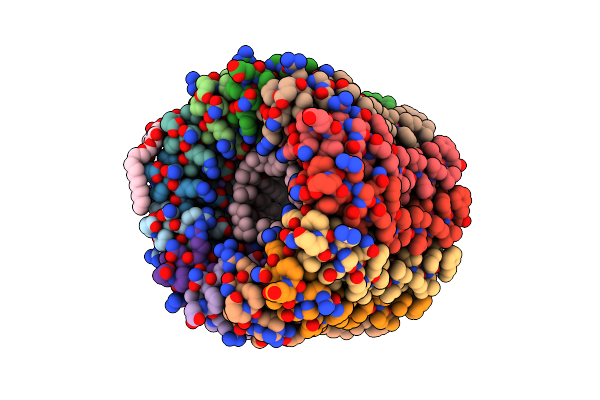
The C-Ring Ion Binding Site Of The Atp Synthase From Bacillus Pseudofirmus Of4 Is Adapted To Alkaliphilic Cell Physiology
Organism: Bacillus pseudofirmus of4
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2014-05-28 Classification: TRANSFERASE Ligands: DPV, TAM, LMT |
 |
The C-Ring Ion Binding Site Of The Atp Synthase From Bacillus Pseudofirmus Of4 Is Adapted To Alkaliphilic Cell Physiology
Organism: Bacillus pseudofirmus of4
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2014-04-23 Classification: TRANSFERASE Ligands: DPV, NA, LMT |
 |
Crystal Structure Of Bacillus Pseudofirmus Of4 Mutant Atp Synthase C12 Ring.
Organism: Bacillus pseudofirmus of4
Method: X-RAY DIFFRACTION Resolution:4.10 Å Release Date: 2013-05-01 Classification: HYDROLASE |
 |
Structural Basis Of A Novel Proton-Coordination Type In An F1Fo-Atp Synthase Rotor Ring
Organism: Bacillus pseudofirmus of4
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2010-08-18 Classification: MEMBRANE PROTEIN Ligands: DPV, NA |

