Search Count: 14
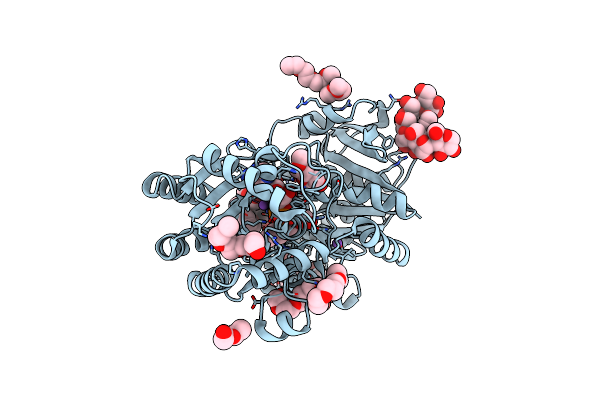 |
Crystal Structure Of E.Coli Gs Mutant E377A In Complex With Adp And Oligosaccharides
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2009-04-28 Classification: TRANSFERASE |
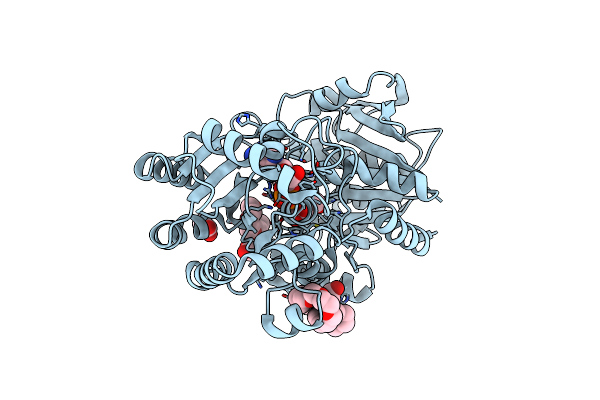 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2009-04-28 Classification: TRANSFERASE Ligands: ADP, 250, ASO, PE3, ACT |
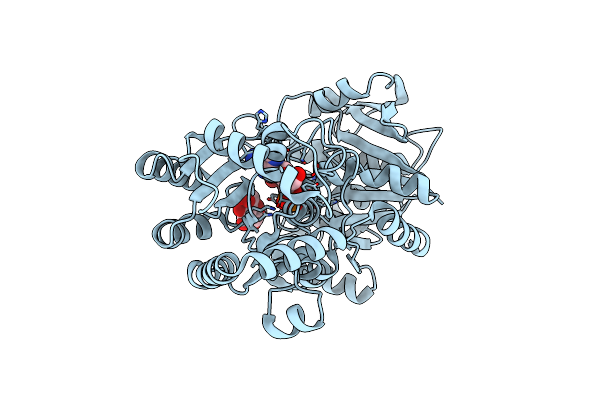 |
Crystal Structure Of E.Coli Gs Mutant E377A In Complex With Adp And Acceptor Analogue Heppso
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2009-03-24 Classification: TRANSFERASE Ligands: GLC, ADP, 250 |
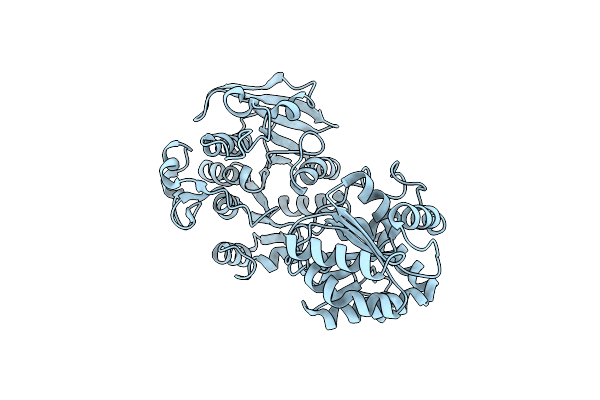 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2009-03-24 Classification: TRANSFERASE |
 |
Crystal Structure Of Wild-Type E.Coli Gs In Complex With Adp And Glucose(Wtgsb)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2008-09-09 Classification: TRANSFERASE Ligands: GLC, ADP, 250 |
 |
Crystal Structure Of Wild-Type E.Coli Gs In Complex With Adp And Glucose(Wtgsc)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2008-09-09 Classification: TRANSFERASE Ligands: GLC, ADP, 250, PE3 |
 |
Crystal Structure Of Wild-Type E.Coli Gs In Complex With Adp And Glucose(Wtgsd)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2008-09-09 Classification: TRANSFERASE Ligands: GLC, ADP, 250, PE3 |
 |
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2006-04-18 Classification: LYASE Ligands: ZN, SO4 |
 |
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2006-04-18 Classification: LYASE Ligands: ZN, BCT, SO4 |
 |
Identification Of A Novel Non-Catalytic Bicarbonate Binding Site In Eubacterial Beta-Carbonic Anhydrase
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2006-04-18 Classification: LYASE Ligands: ZN, BCT |
 |
Organism: Solanum tuberosum
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2005-03-15 Classification: TRANSFERASE Ligands: SO4, PMB |
 |
Crystal Structure Of Potato Tuber Adp-Glucose Pyrophosphorylase In Complex With Atp
Organism: Solanum tuberosum
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2005-03-15 Classification: TRANSFERASE Ligands: SO4, ATP |
 |
Crystal Structure Of Potato Tuber Adp-Glucose Pyrophosphorylase In Complex With Adp-Glucose
Organism: Solanum tuberosum
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2005-03-15 Classification: TRANSFERASE Ligands: SO4, ADP, ADQ |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2002-09-18 Classification: TRANSFERASE |

