Search Count: 4
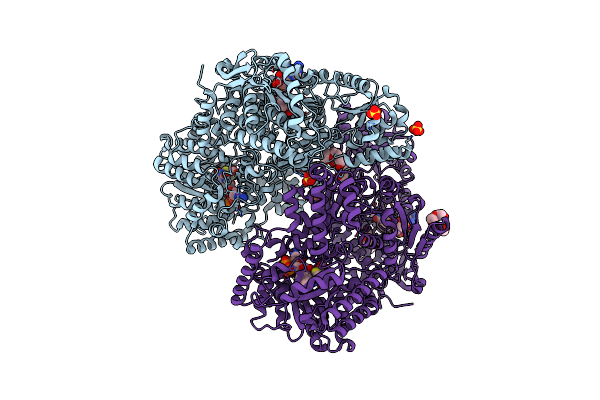 |
Structure Of Proline Utilization A With 1,3-Dithiolane-2-Carboxylate Bound In The Proline Dehydrogenase Active Site
Organism: Sinorhizobium meliloti (strain sm11)
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2021-09-29 Classification: OXIDOREDUCTASE Ligands: UJD, FAD, NAD, SO4, MG, PGE, PEG, PG4 |
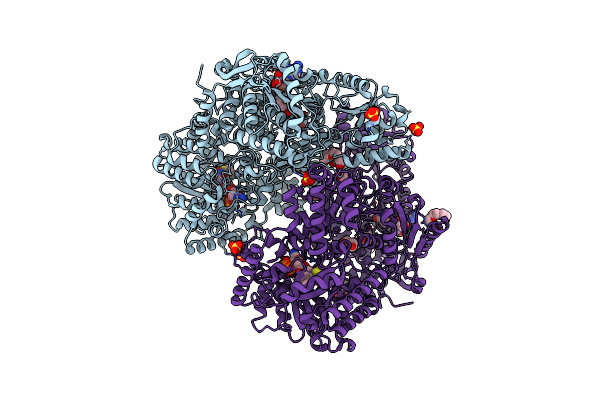 |
Structure Of Proline Utilization A With The Fad Covalently-Modified By 1,3-Dithiolane
Organism: Sinorhizobium meliloti sm11
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2021-09-29 Classification: OXIDOREDUCTASE Ligands: UJJ, MG, NAD, SO4, PGE, PEG, PG4 |
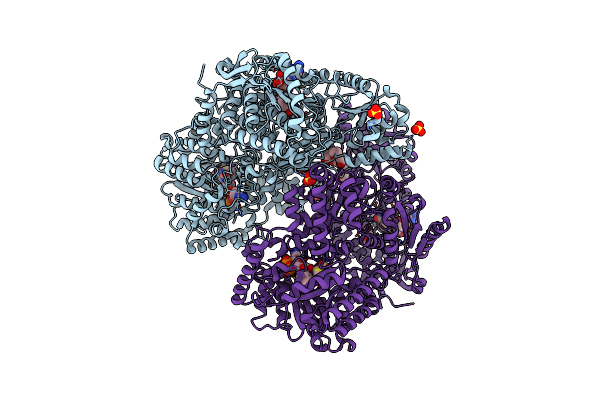 |
Structure Of Proline Utilization A With Tetrahydrothiophene-2-Carboxylate Bound In The Proline Dehydrogenase Active Site
Organism: Sinorhizobium meliloti (strain sm11)
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2021-09-29 Classification: OXIDOREDUCTASE Ligands: UJM, UJP, PGE, FAD, NAD, SO4, MG, PEG |
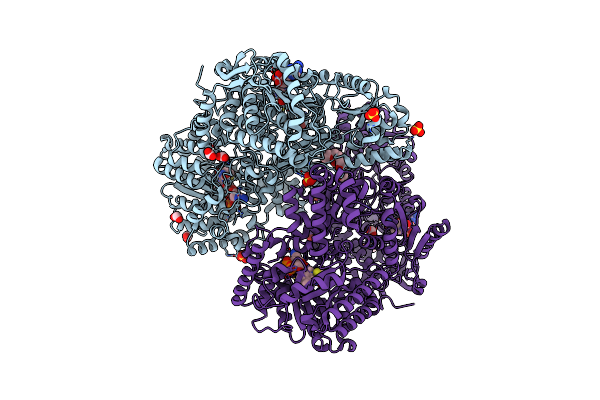 |
Structure Of Proline Utilization A With The Fad Covalently Modified By Tetrahydrothiophene
Organism: Sinorhizobium meliloti sm11
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2021-09-29 Classification: OXIDOREDUCTASE Ligands: UJG, NAI, SO4, PGE, FMT, MG, PEG |

