Search Count: 36
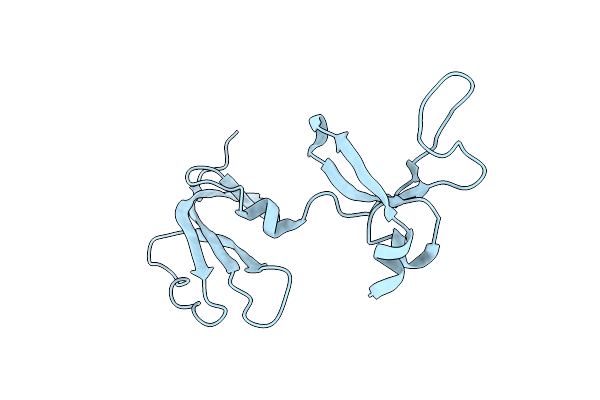 |
Crystal Structure Of Pilin-Specific Sortase Srtc2 From Lactobacillus Rhamnosus Gg
Organism: Lacticaseibacillus rhamnosus gg
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2023-08-30 Classification: HYDROLASE |
 |
E. Faecium Muraa In Complex With Udp-N-Acetylmuramic Acid (Unam) And A Covalent Adduct Of Pep With Cys119
Organism: Enterococcus faecalis
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2023-07-05 Classification: TRANSFERASE Ligands: EPZ |
 |
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2022-12-28 Classification: TRANSFERASE Ligands: UD1, FFQ, K, NA, CL |
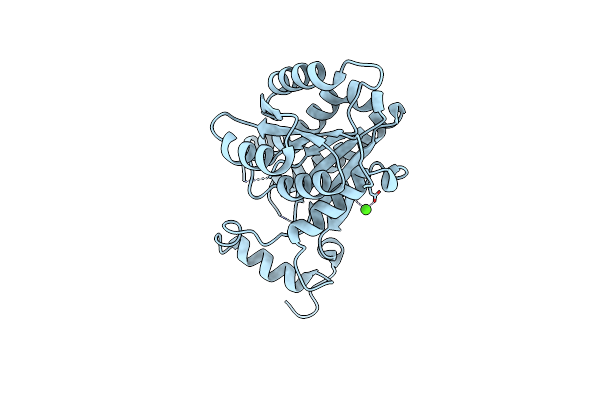 |
Crystal Structure Of The Apo Enoyl-Acp-Reductase (Fabi) From Moraxella Catarrhalis
Organism: Moraxella catarrhalis (strain bbh18)
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2022-07-20 Classification: OXIDOREDUCTASE Ligands: CA |
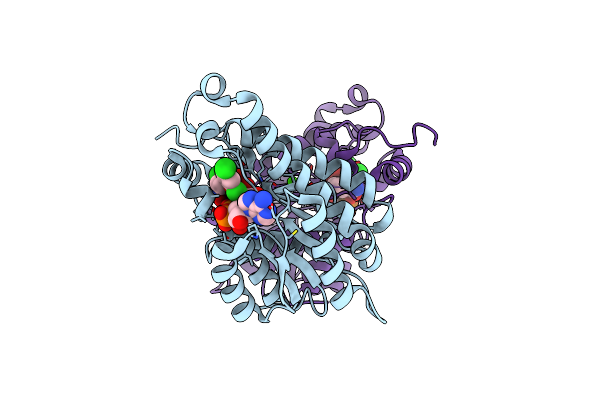 |
Crystal Structure Of Moraxella Catarrhalis Enoyl-Acp-Reductase (Fabi) In Complex With Nad And Triclosan
Organism: Moraxella catarrhalis (strain bbh18)
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2022-07-20 Classification: OXIDOREDUCTASE Ligands: NAD, TCL, CA |
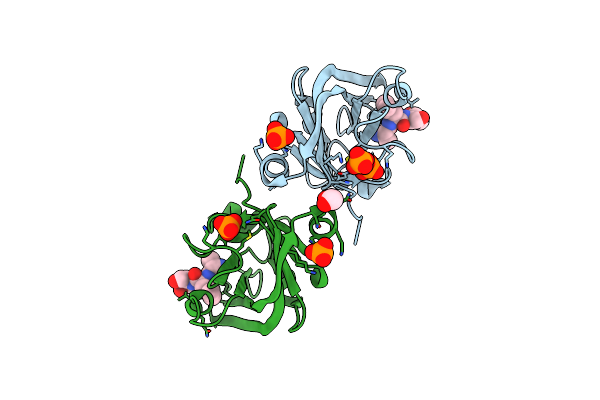 |
Crystal Structure Of Housekeeping Sortase Srta Bound With Self Derived Tripeptide From Lactobacillus Rhamnosus Gg
Organism: Lactobacillus rhamnosus, Lacticaseibacillus rhamnosus
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2022-07-06 Classification: TRANSFERASE Ligands: PO4, EDO |
 |
Crystal Structure Of Moraxella Catarrhalis Enoyl-Acp-Reductase (Fabi) In Complex With The Cofactor Nad
Organism: Moraxella catarrhalis (strain bbh18)
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2022-06-22 Classification: OXIDOREDUCTASE Ligands: NAD, CA, GOL |
 |
Crystal Structure Of Type Iii Polyketide Synthase From Mycobacterium Marinum
Organism: Mycobacterium marinum (strain atcc baa-535 / m)
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2022-06-15 Classification: TRANSFERASE Ligands: MYR, PGE, ACT |
 |
Crystal Structure Of Type Iii Polyketide Synthase From Mycobacterium Marinum - P 21 21 21 Space Group
Organism: Mycobacterium marinum (strain atcc baa-535 / m)
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2022-03-23 Classification: TRANSFERASE Ligands: OCA |
 |
Crystal Structure Of Housekeeping Sortase Srta From Lactobacillus Rhamnosus Gg
Organism: Lactobacillus rhamnosus
Method: X-RAY DIFFRACTION Resolution:2.64 Å Release Date: 2022-03-02 Classification: HYDROLASE |
 |
Crystal Structure Of Pilin-Specific Sortase Srtc1 From Lactobacillus Rhamnosus Gg
Organism: Lactobacillus rhamnosus
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2022-03-02 Classification: HYDROLASE Ligands: SO4 |
 |
X-Ray Crystal Structure Of Vapb12 Antitoxin From Mycobacterium Tuberculosis In Space Group P41.
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2022-02-16 Classification: ANTITOXIN Ligands: ZN |
 |
Organism: Lactobacillus rhamnosus
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2020-12-30 Classification: CELL ADHESION Ligands: MG, CL |
 |
Organism: Momordica charantia
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2020-02-12 Classification: PLANT PROTEIN Ligands: CU, ACT |
 |
Organism: lactobacillus rhamnosus gg
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2018-06-20 Classification: CELL ADHESION |
 |
Crystal Structure Of C-Terminal Fragment Of Spad From Lactobacillus Rhamnosus Gg Generated By Limited Proteolysis
Organism: Lactobacillus rhamnosus gg
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2018-06-20 Classification: CELL ADHESION Ligands: CL |
 |
Crystal Structure Of Shaft Pilin Spad From Lactobacillus Rhamnosus Gg In Bent Conformation
Organism: Lactobacillus rhamnosus gg
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2018-06-20 Classification: CELL ADHESION |
 |
Crystal Structure Of Shaft Pilin Spad From Lactobacillus Rhamnosus Gg - D242A Mutant
Organism: lactobacillus rhamnosus gg
Method: X-RAY DIFFRACTION Resolution:2.47 Å Release Date: 2018-06-20 Classification: CELL ADHESION |
 |
Crystal Structure Of Shaft Pilin Spad From Lactobacillus Rhamnosus Gg - K365A Mutant
Organism: Lactobacillus rhamnosus gg
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2018-06-20 Classification: CELL ADHESION |
 |
Crystal Structure Of Chorismate Mutase Like Domain Of Bifunctional Dahp Synthase Of Bacillus Subtilis In Complex With Chlorogenic Acid
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-07-26 Classification: ISOMERASE Ligands: 7LH, SO4 |

