Search Count: 61
 |
Organism: Homo sapiens, Xenopus laevis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: CELL CYCLE |
 |
Organism: Synthetic construct, Xenopus laevis, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: CELL CYCLE |
 |
Organism: Xenopus laevis, Synthetic construct, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: CELL CYCLE |
 |
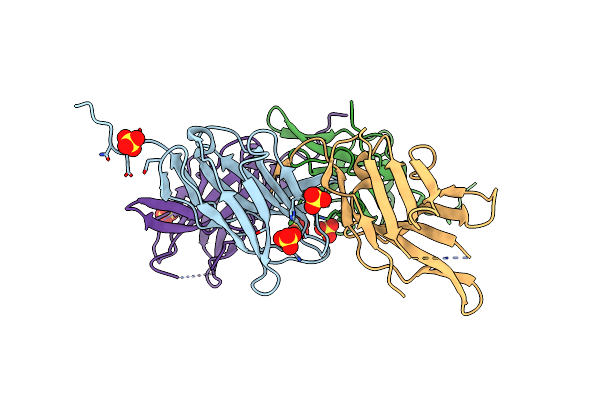
Chromosomal Passenger Complex In Complex With H3T3Ph Nucleosome (Double Occupancy)
Organism: Homo sapiens, Xenopus laevis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: CELL CYCLE |
 |
Organism: Oryza, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: DNA BINDING PROTEIN |
 |
Organism: Oryza, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: DNA BINDING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2024-10-23 Classification: CELL CYCLE Ligands: SAH |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.62 Å Release Date: 2024-10-23 Classification: CELL CYCLE Ligands: SAM |
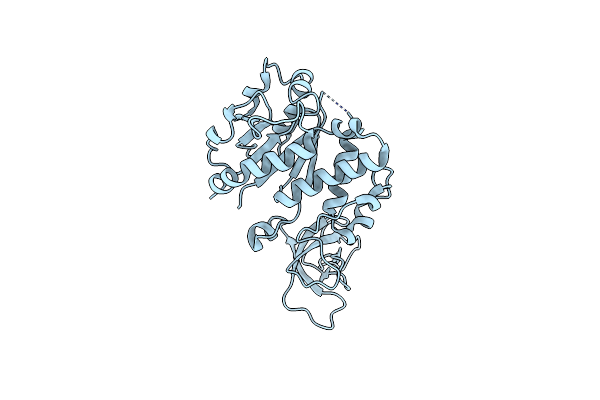 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-10-23 Classification: CELL CYCLE Ligands: HOH |
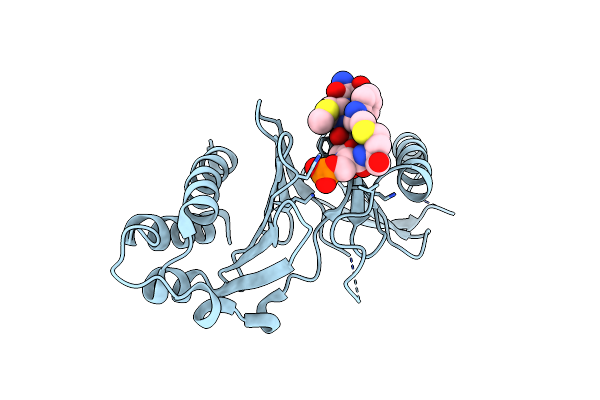 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2024-08-21 Classification: CELL CYCLE |
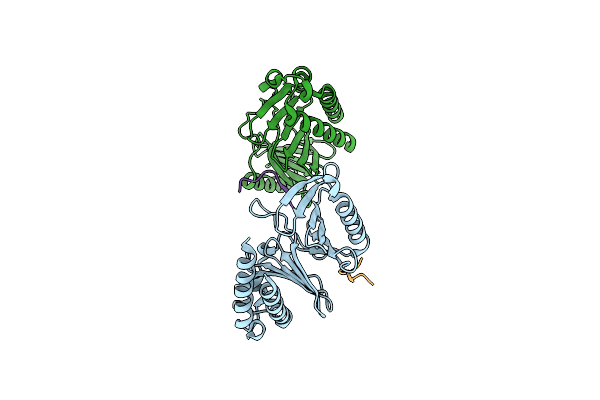 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2024-08-21 Classification: CELL CYCLE |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2023-10-11 Classification: PROTEIN BINDING Ligands: SO4 |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-02-01 Classification: TRANSFERASE Ligands: MLA |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-02-01 Classification: TRANSFERASE Ligands: ACT, MLA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-03-02 Classification: HYDROLASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2022-03-02 Classification: HYDROLASE Ligands: ATP, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2022-03-02 Classification: HYDROLASE Ligands: ATP, MG |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:4.36 Å Release Date: 2020-04-15 Classification: CELL CYCLE |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:3.47 Å Release Date: 2020-04-01 Classification: CELL CYCLE |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2020-04-01 Classification: CELL CYCLE |

