Search Count: 19
 |
Crystal Structure Of The Hect Domain Of Smurf 2 In Complex With Inhibitor 8
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2025-04-16 Classification: LIGASE Ligands: TJQ, PO4 |
 |
Crystal Structure Of The Hect Domain Of Smurf 1 In Complex With Inhibitor 8
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2025-04-16 Classification: LIGASE Ligands: TJQ |
 |
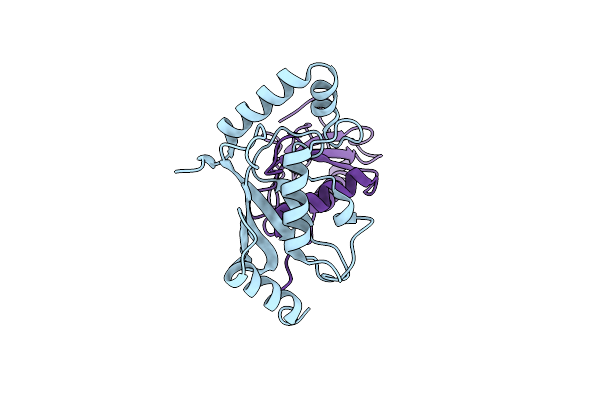
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2025-04-16 Classification: LIGASE |
 |
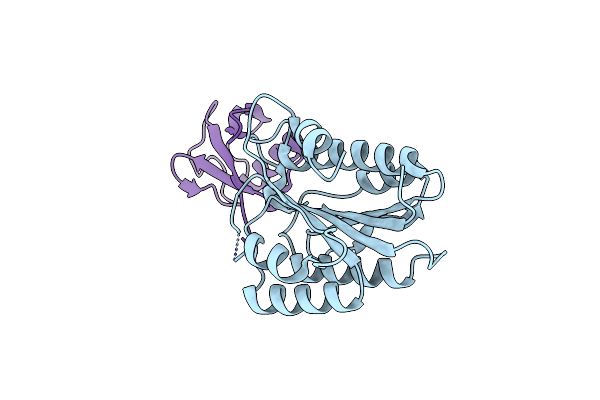
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-08-02 Classification: SIGNALING PROTEIN |
 |
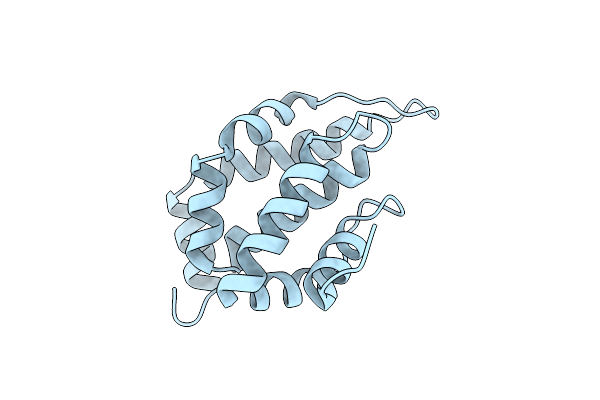
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.14 Å Release Date: 2016-10-19 Classification: UBIQUITIN-BINDING PROTEIN |
 |
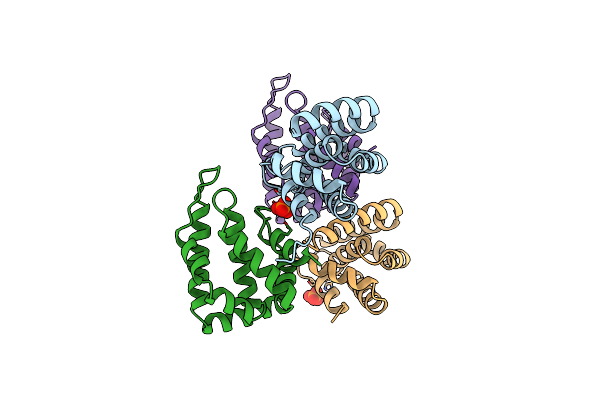
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2016-10-05 Classification: UBIQUITIN-BINDING DOMAIN |
 |
Crystal Structure Of The Zebra Fish Enth Domain From Epsin1 In 1.41 Angstrom Resolution
Organism: Danio rerio
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2016-10-05 Classification: SIGNALING PROTEIN Ligands: PO4 |
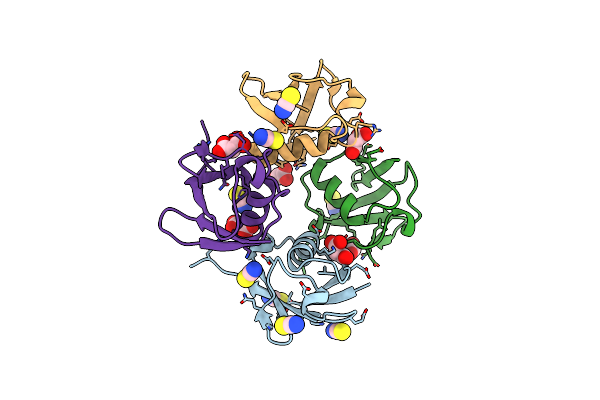 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2015-07-29 Classification: CELL CYCLE Ligands: MLA, SCN |
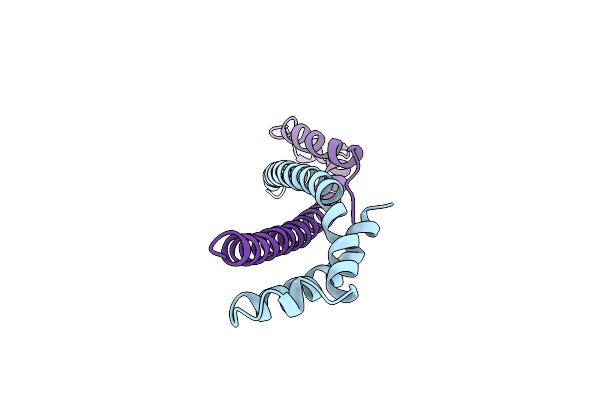 |
The Vps27/Hse1 Complex Is A Gat Domain-Based Scaffold For Ubiquitin-Dependent Sorting
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.01 Å Release Date: 2007-06-05 Classification: ENDOCYTOSIS/EXOCYTOSIS |
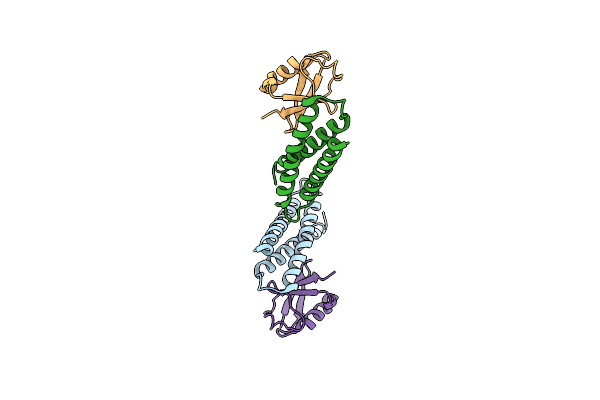 |
Organism: Homo sapiens, Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2005-02-22 Classification: PROTEIN TRANSPORT, CHROMOSOMAL PROTEIN |
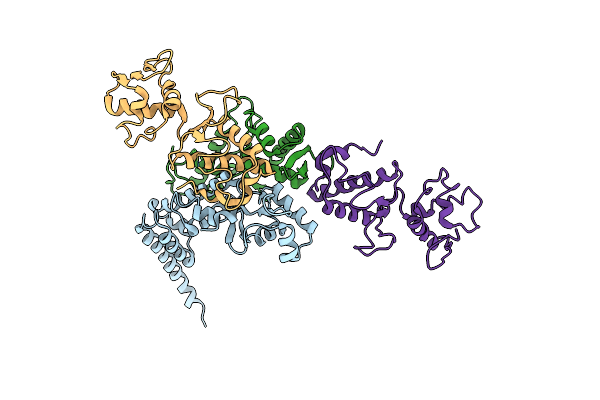 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2004-09-21 Classification: TRANSPORT PROTEIN |
 |
Organism: Saccharomyces cerevisiae, Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2003-06-24 Classification: TRANSLATION |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2003-06-10 Classification: PROTEIN TRANSPORT |
 |
Organism: Serratia marcescens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2002-11-06 Classification: HYDROLASE |
 |
Crystal Structure Of Chitinase A Mutant Y390F Complexed With Hexa-N-Acetylchitohexaose (Nag)6
Organism: Serratia marcescens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2001-09-26 Classification: HYDROLASE |
 |
Crystal Structure Of Chitinase A Mutant D313A Complexed With Octa-N-Acetylchitooctaose (Nag)8.
Organism: Serratia marcescens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2001-02-25 Classification: HYDROLASE |
 |
Crystal Structure Of Chitinase A Mutant E315Q Complexed With Octa-N-Acetylchitooctaose (Nag)8.
Organism: Serratia marcescens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2001-02-22 Classification: HYDROLASE |
 |
Beta-N-Acetylhexosaminidase Mutant D539A Complexed With Di-N-Acetyl-Beta-D-Glucosamine (Chitobiase)
Organism: Serratia marcescens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2000-09-20 Classification: HYDROLASE Ligands: SO4 |
 |
Beta-N-Acetylhexosaminidase Mutant E540D Complexed With Di-N Acetyl-D-Glucosamine (Chitobiase)
Organism: Serratia marcescens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2000-09-20 Classification: HYDROLASE Ligands: SO4 |

