Search Count: 205
 |
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2025-10-22 Classification: OXIDOREDUCTASE Ligands: NDP, A1IXO, EDO |
 |
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2025-10-22 Classification: OXIDOREDUCTASE Ligands: NDP, A1IXP, EDO |
 |
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2025-10-22 Classification: OXIDOREDUCTASE Ligands: NDP, A1IXQ, ACT, EDO, CL |
 |
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2025-10-22 Classification: OXIDOREDUCTASE Ligands: NDP, A1IXS, ACT, EDO |
 |
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-10-22 Classification: OXIDOREDUCTASE Ligands: NDP, A1IXR, EDO |
 |
Sc-4E (Scfv Derived From The Mab 4E1 Against Cd93) Crystallized At Ph 7.5 (Dimeric State From Gel Filtration)
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2025-04-30 Classification: IMMUNE SYSTEM Ligands: CL, EDO, IMD, NA |
 |
Sc-4E (Scfv Derived From The Mab 4E1 Against Cd93) Crystallized At Ph 7.5 (Monomeric State From Gel Filtration)
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-04-30 Classification: IMMUNE SYSTEM Ligands: EDO, IMD, NA, CL |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2025-04-30 Classification: IMMUNE SYSTEM Ligands: CL, EDO, IMD, NA |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-04-30 Classification: IMMUNE SYSTEM Ligands: CL, EDO, IMD, NA |
 |
M2 Mutant (R111K:Y134F:T54V:R132Q:P39Y:R59Y) Of Human Cellular Retinoic Acid Binding Protein Ii - 1E Conjugate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-03-26 Classification: TRANSPORT PROTEIN Ligands: A1IQZ |
 |
M2 Mutant (R111K:Y134F:T54V:R132Q:P39Y:R59Y) Of Human Cellular Retinoic Acid Binding Protein Ii - 1M Conjugate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2025-03-26 Classification: TRANSPORT PROTEIN Ligands: A1IQY |
 |
M2 Mutant (R111K:Y134F:T54V:R132Q:P39Y:R59Y) Of Human Cellular Retinoic Acid Binding Protein Ii - 1J Conjugate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2025-03-26 Classification: TRANSPORT PROTEIN Ligands: A1IQ0 |
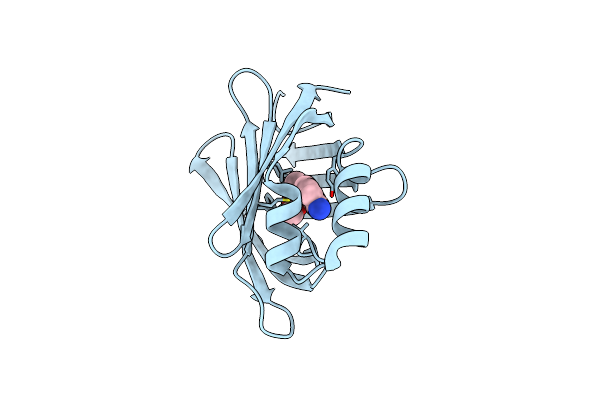 |
M2 Mutant (R111K:Y134F:T54V:R132Q:P39Y:R59Y) Of Human Cellular Retinoic Acid Binding Protein Ii - 1L Conjugate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2025-03-26 Classification: TRANSPORT PROTEIN Ligands: A1IQ1 |
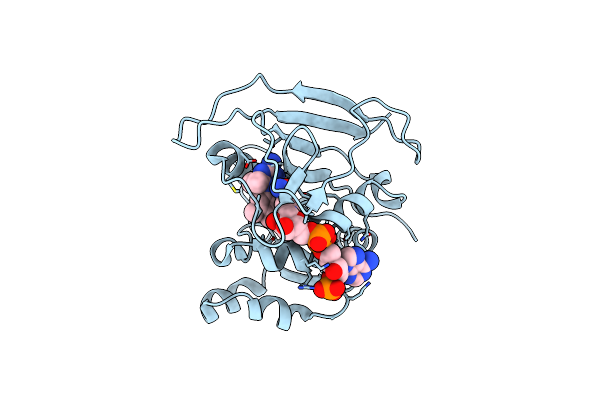 |
Crystal Structure Of Trypanosoma Brucei Dhfr In Complex With The Cofactor And Inhibitor P25
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2024-11-20 Classification: OXIDOREDUCTASE Ligands: NDP, A1H0S |
 |
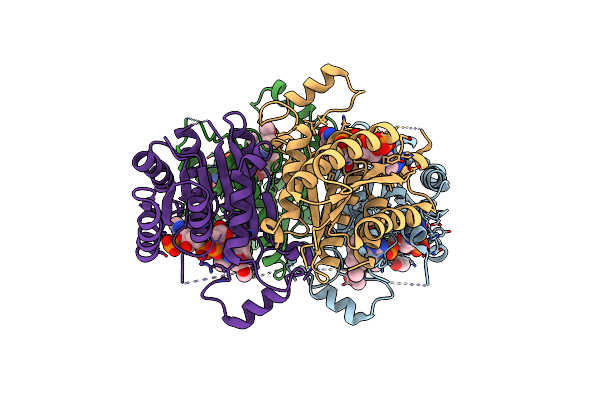
Crystal Structure Of Trypanosoma Brucei Ptr1 In Complex With The Cofactor And Inhibitor P25
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2024-11-20 Classification: OXIDOREDUCTASE Ligands: NDP, A1H0S, ACT |
 |
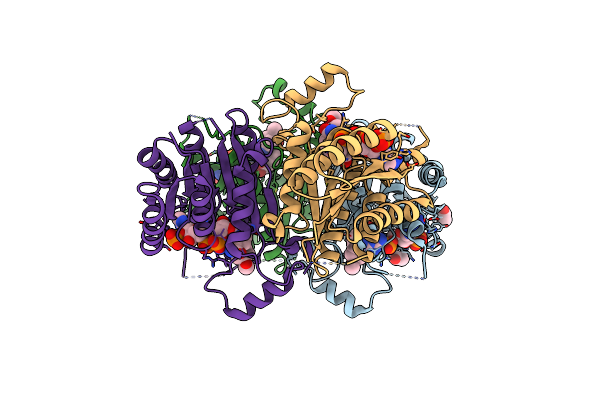
Crystal Structure Of Trypanosoma Brucei Ptr1 In Complex With The Cofactor And Inhibitor P30
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-11-20 Classification: OXIDOREDUCTASE Ligands: NDP, ACT, A1H0U |
 |
Crystal Structure Of Trypanosoma Brucei Ptr1 In Complex With The Cofactor And Inhibitor P31
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-11-20 Classification: OXIDOREDUCTASE Ligands: NDP, A1H0W, ACT |
 |
Crystal Structure Of Trypanosoma Brucei Ptr1 In Complex With The Cofactor And Inhibitor P32
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2024-11-20 Classification: OXIDOREDUCTASE Ligands: NDP, A1H0T, ACT |
 |
Crystal Structure Of Trypanosoma Brucei Ptr1 In Complex With The Cofactor And Inhibitor P34
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2024-11-20 Classification: OXIDOREDUCTASE Ligands: NDP, A1H0V |
 |
Crystal Structure Of Recombinant Chicken Liver Bile Acid Binding Protein (Cl-Babp) In Complex With Chenodeoxycholic Acid
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-10-09 Classification: LIPID BINDING PROTEIN Ligands: JN3 |

