Search Count: 44
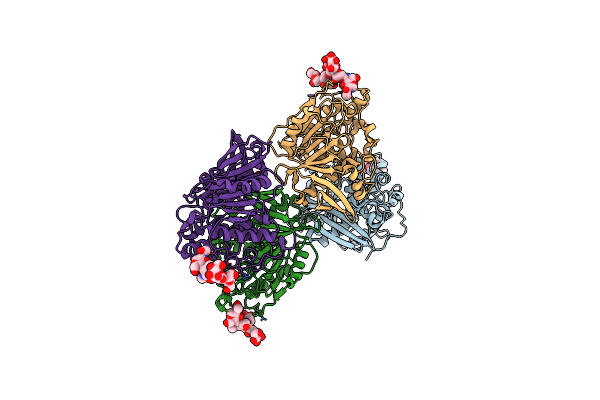 |
Structure Of Lentithecium Fluviatile Carbohydrate Esterase From The Ce15 Family (Lfce15C)
Organism: Lentithecium fluviatile
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2023-06-07 Classification: HYDROLASE Ligands: FMT |
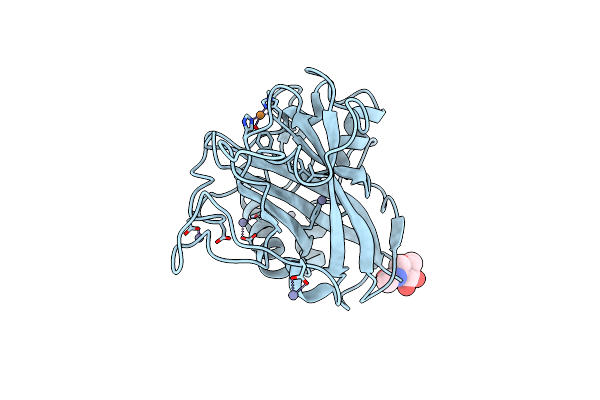 |
Aa13 Lytic Polysaccharide Monooxygenase From Aspergillus Oryzae Partially In Cu(Ii) State
Organism: Aspergillus oryzae rib40
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2020-03-18 Classification: OXIDOREDUCTASE Ligands: CU, NAG, ZN |
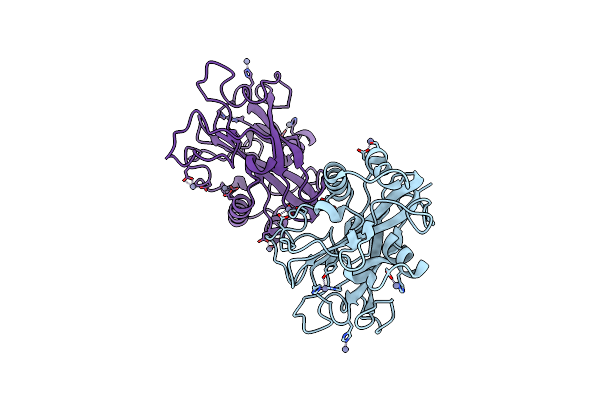 |
Glycosylated Aa13 Lytic Polysaccharide Monooxygenase From Aspergillus Oryzae In P1 Space Group
Organism: Aspergillus oryzae rib40
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-03-18 Classification: OXIDOREDUCTASE Ligands: ZN |
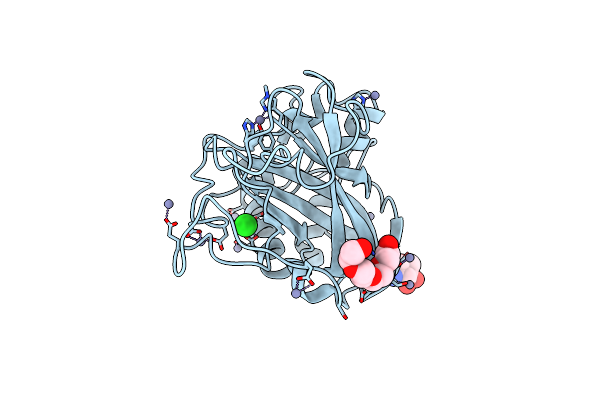 |
Aa13 Lytic Polysaccharide Monooxygenase From Aspergillus Oryzae Measured With Ssx
Organism: Aspergillus oryzae rib40
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-03-18 Classification: METAL BINDING PROTEIN Ligands: NAG, ZN, CL, PG4 |
 |
Organism: Aspergillus aculeatus
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2019-06-12 Classification: HYDROLASE Ligands: PEG, CA, PG4, EDO, ACT, PG6, PGE |
 |
Organism: Lentinus similis
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2017-11-01 Classification: UNKNOWN FUNCTION Ligands: CU, NAG, CL, BGC, BMA |
 |
Organism: Lentinus similis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-11-01 Classification: OXIDOREDUCTASE Ligands: CU, NAG, CL, XYP |
 |
Organism: Lentinus similis
Method: X-RAY DIFFRACTION Resolution:1.33 Å Release Date: 2017-11-01 Classification: OXIDOREDUCTASE Ligands: CU, NAG, CL |
 |
Organism: Lentinus similis
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2017-11-01 Classification: OXIDOREDUCTASE Ligands: CU, NAG, XYP |
 |
Organism: Lentinus similis
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2017-11-01 Classification: OXIDOREDUCTASE Ligands: CU, NAG, XYP, CL |
 |
Organism: Lentinus similis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-11-01 Classification: OXIDOREDUCTASE Ligands: CU, NAG, CL |
 |
Organism: Lentinus similis
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2017-11-01 Classification: OXIDOREDUCTASE Ligands: CU, NAG, CL, NA |
 |
Organism: Collariella virescens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-11-01 Classification: OXIDOREDUCTASE Ligands: CU, SO4 |
 |
Organism: Aspergillus oryzae rib40
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2017-01-18 Classification: METAL BINDING PROTEIN Ligands: NAG, ZN |
 |
Organism: Aspergillus oryzae rib40
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2017-01-18 Classification: METAL BINDING PROTEIN Ligands: NAG, ZN |
 |
Organism: Aspergillus oryzae rib40
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2017-01-18 Classification: METAL BINDING PROTEIN Ligands: NAG, ZN |
 |
Organism: Aspergillus oryzae rib40
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2017-01-11 Classification: METAL BINDING PROTEIN Ligands: NAG, ZN |
 |
Structural Characterization Of The Thermostable Bradyrhizobium Japonicum D-Sorbitol Dehydrogenase
Organism: Bradyrhizobium japonicum
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2016-11-09 Classification: OXIDOREDUCTASE Ligands: PO4, SOR |
 |
Organism: Lentinus similis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-03-02 Classification: OXIDOREDUCTASE Ligands: CU, NAG, CL |
 |
Organism: Lentinus similis
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2016-03-02 Classification: OXIDOREDUCTASE Ligands: CU, NAG, CL |

