Search Count: 9
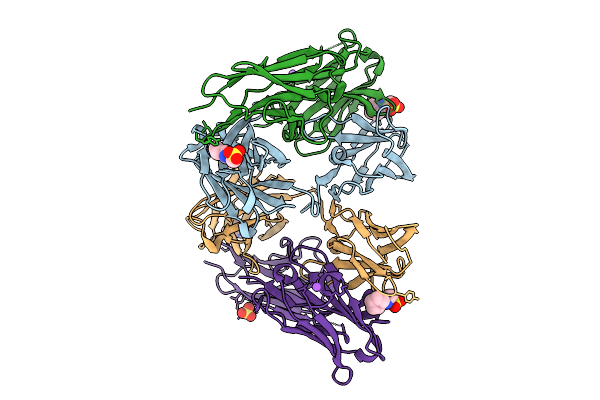 |
Crystal Structure Of A High Affinity Vl-Vh Tetrabody For The Erythropoietin Receptor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: SIGNALING PROTEIN Ligands: NHE, CL, SO4, NA |
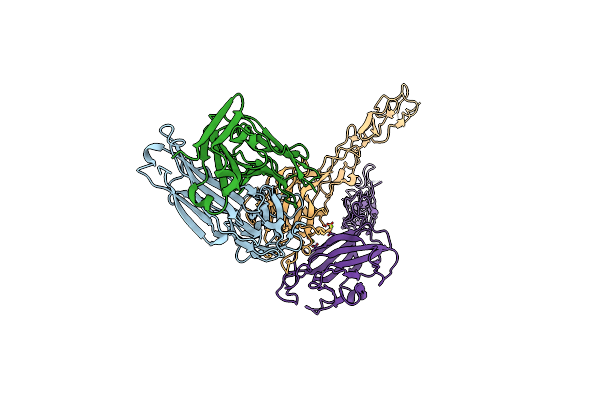 |
Structure Of The Epha2 Lbdcrd Bound To Fabs1Ce_L1 In A 2:1 (Epha2 To Fab) Ratio
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2024-09-11 Classification: SIGNALING PROTEIN/IMMUNE SYSTEM Ligands: MG |
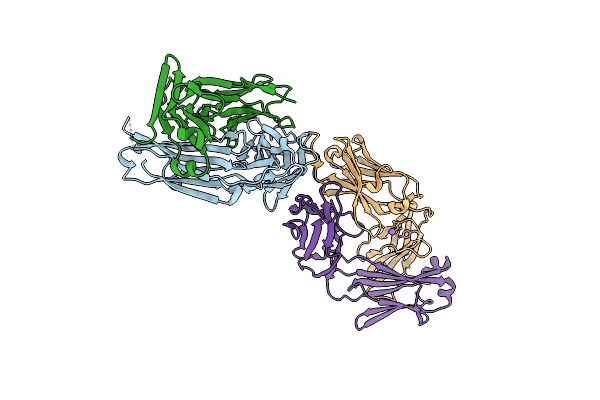 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2024-07-31 Classification: SIGNALING PROTEIN/IMMUNE SYSTEM Ligands: CL, NA |
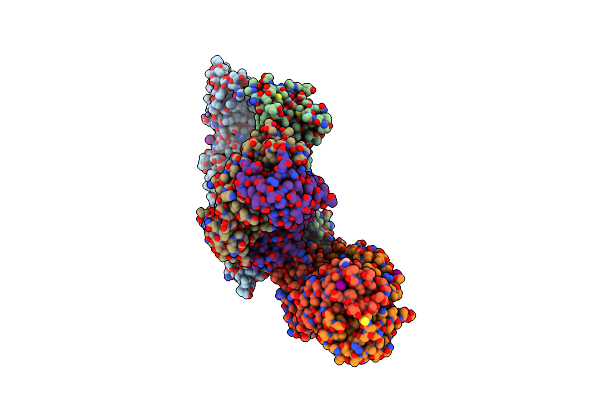 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2024-07-17 Classification: SIGNALING PROTEIN/IMMUNE SYSTEM Ligands: IOD, CL, NA, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2024-06-26 Classification: SIGNALING PROTEIN/IMMUNE SYSTEM Ligands: CL, EDO |
 |
Structure Of The Ck Variant Of Fab F1 (Fabc-F1) In Complex With The C-Terminal Fn3 Domain Of Epha2
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2024-05-01 Classification: IMMUNE SYSTEM |
 |
 |
Hydrogen Bonding And Catalysis: An Unexpected Explanation For How A Single Amino Acid Substitution Can Change The Ph Optimum Of A Glycosidase
Organism: Bacillus circulans
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2000-05-12 Classification: HYDROLASE |
 |
Hydrogen Bonding And Catalysis: An Unexpected Explanation For How A Single Amino Acid Substitution Can Change The Ph Optimum Of A Glycosidase
Organism: Bacillus circulans
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2000-05-12 Classification: HYDROLASE |

