Search Count: 192
 |
Lh2 Complex From Ectothiorhodospira Haloalkaliphila With Inhibited Carotenoid Biosynthesis
Organism: Ectothiorhodospira haloalkaliphila atcc 51935
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: PHOTOSYNTHESIS Ligands: BCL, A1MBA |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: TRANSCRIPTION |
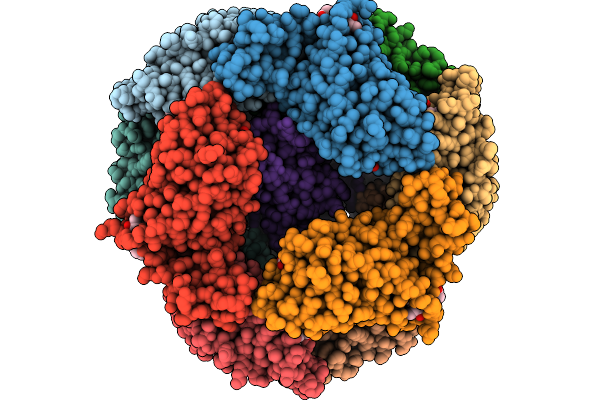 |
Organism: Gloeobacter violaceus pcc 7421
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: PHOTOSYNTHESIS Ligands: CYC |
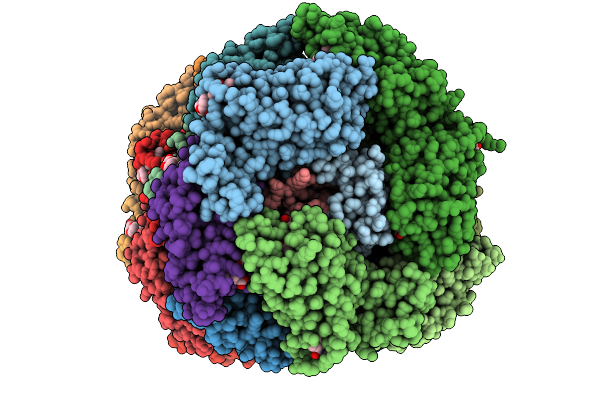 |
Organism: Gloeobacter violaceus pcc 7421
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: PHOTOSYNTHESIS Ligands: CYC |
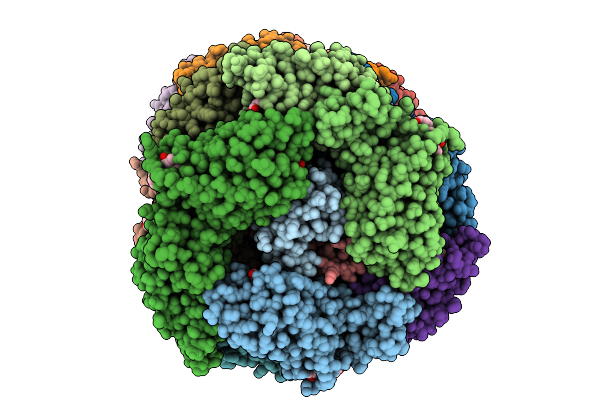 |
Organism: Gloeobacter violaceus pcc 7421
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: PHOTOSYNTHESIS Ligands: CYC |
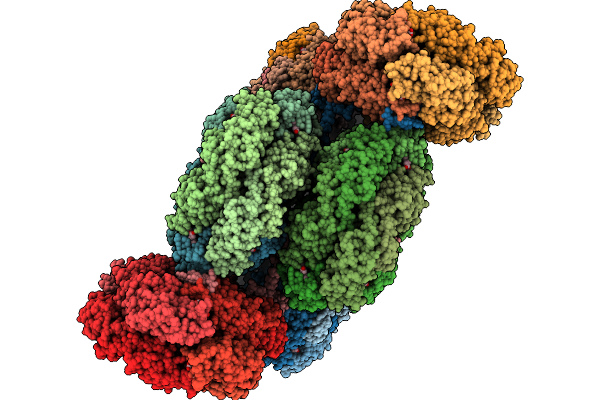 |
Organism: Gloeobacter violaceus pcc 7421
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: PHOTOSYNTHESIS Ligands: CYC |
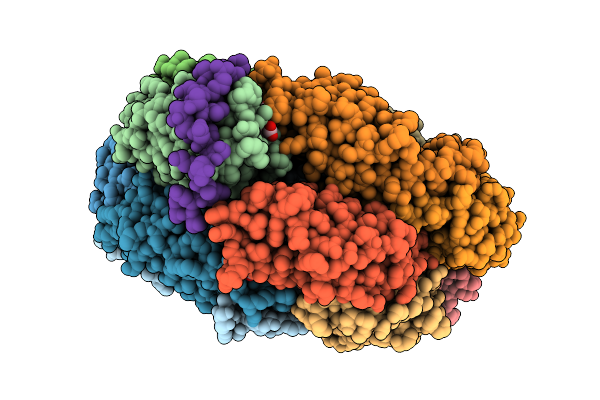 |
Phycobilisome Allophycocyanin Hexamer C From Gloeobacter Violaceus Pcc 7421
Organism: Gloeobacter violaceus pcc 7421
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: PHOTOSYNTHESIS Ligands: CYC |
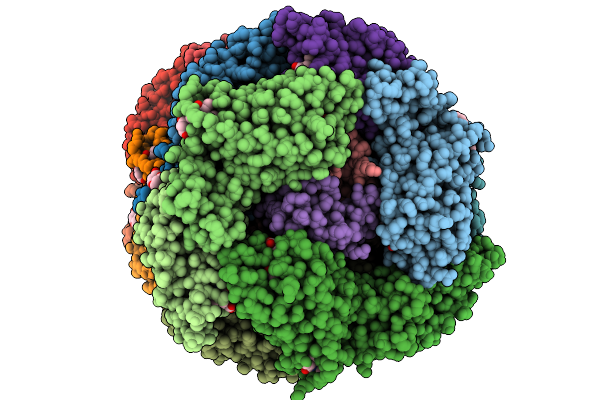 |
Organism: Gloeobacter violaceus pcc 7421
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: PHOTOSYNTHESIS Ligands: CYC |
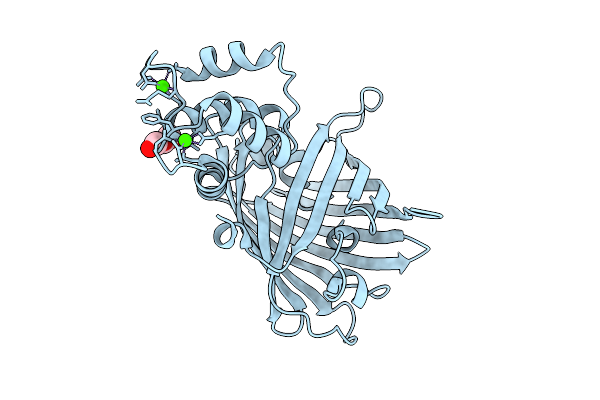 |
Crystal Structure Of The Genetically-Encoded Green Calcium Sensor Icbtnc2 In Its Calcium Bound-State
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: FLUORESCENT PROTEIN Ligands: PEG, CA, CL |
 |
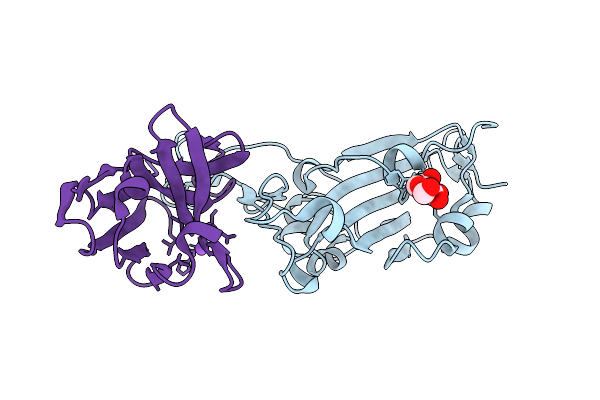
Nmr Solution Structure Of Termini-Truncated Oxidized Cytochrome C552 From Thioalkalivibrio Paradoxus
Organism: Thioalkalivibrio paradoxus arh 1
Method: SOLUTION NMR Release Date: 2025-07-30 Classification: ELECTRON TRANSPORT Ligands: HEC |
 |
Crystal Structure Of The Wuhan Sars-Cov-2 Spike Rbd (319-541) Complexed With 1P1B10 Nanobody
Organism: Severe acute respiratory syndrome coronavirus 2, Camelus bactrianus
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG, NA |
 |
Crystal Structure Of The Soluble Green Pigment Protein From Tettigonia Cantans
Organism: Tettigonia cantans
Method: X-RAY DIFFRACTION Release Date: 2025-04-23 Classification: TRANSPORT PROTEIN Ligands: LUT, AZI, PLC, A1L6M |
 |
Crystal Structure L-Lactate Dehydrogenase From Lactobacillus Reuteri In Its Apoform
Organism: Limosilactobacillus reuteri
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2025-04-02 Classification: OXIDOREDUCTASE Ligands: EDO |
 |
Crystal Structure Of Aminotransferase-Like Protein From Variovorax Paradoxus Mutant N174K
Organism: Variovorax paradoxus b4
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-01-29 Classification: TRANSFERASE Ligands: PEG |
 |
Lh2 Complex From Ectothiorhodospira Haloalkaliphila At Near-Atomic Resolution
Organism: Ectothiorhodospira haloalkaliphila atcc 51935
Method: ELECTRON MICROSCOPY Resolution:1.70 Å Release Date: 2025-01-01 Classification: PHOTOSYNTHESIS Ligands: BCL, A1L0S |
 |
Crystal Structure Of The C. Difficile Toxin A Crops Domain Fragment 2592-2710 Bound To H5.2 Nanobody
Organism: Camelus dromedarius, Clostridioides difficile 342
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-12-18 Classification: TOXIN/IMMUNE SYSTEM Ligands: EDO, PO4 |
 |
Crystal Structure Of The C. Difficile Toxin A Crops Domain Fragment 2639-2707 Bound To C4.2 Nanobody
Organism: Camelus dromedarius, Clostridioides difficile
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-12-18 Classification: TOXIN/IMMUNE SYSTEM Ligands: EDO |
 |
Crystal Structure Of Cysteine Synthase A From Limosilactobacillus Reuteri Lr1 In Its Apo Form
Organism: Limosilactobacillus reuteri
Method: X-RAY DIFFRACTION Release Date: 2024-11-13 Classification: TRANSFERASE Ligands: PGE, EDO, PEG, SO4 |
 |
Crystal Structure Of Beta-Carotene-Binding Protein (Bbp) From Schistocerca Gregaria Complexed With Beta-Carotene
Organism: Schistocerca gregaria
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2024-09-18 Classification: LIPID BINDING PROTEIN Ligands: BCR |
 |
Crystal Structure Of Monomeric Plp-Dependent Transaminase From Desulfobacula Toluolica In F 41 3 2 Space Group
Organism: Desulfobacula toluolica
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2024-09-18 Classification: TRANSFERASE |

