Maintenance Mode: Some services may turn out to be unavailable. We apologize for the inconvenience!
Maintenance Mode: Some services may turn out to be unavailable. We apologize for the inconvenience!
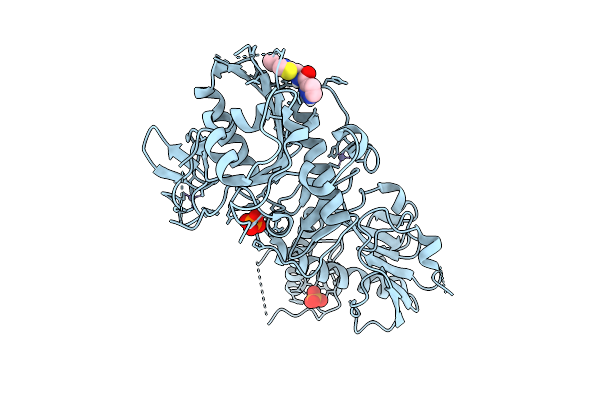 |
Structure Of Sars-Cov-2 Nsp14 Bound To N-((4-Vinylthiazol-2-Yl)Methyl)-1H-Pyrazole-3-Carboxamide
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: VIRAL PROTEIN Ligands: ZN, PO4, A1BX0 |
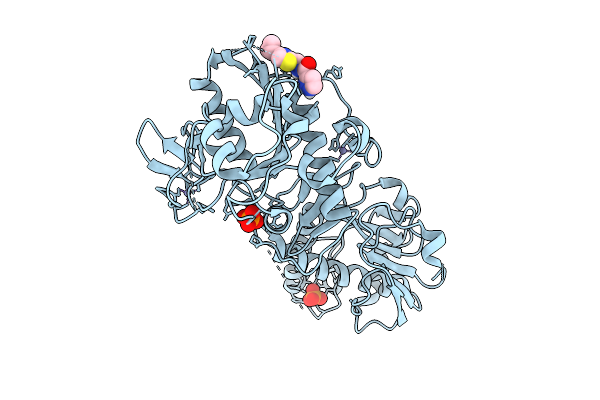 |
Structure Of Sars-Cov-2 Nsp14 Bound To N-((4-Cyclopropylthiazol-2-Yl)Methyl)-1H-Pyrazole-3-Carboxamide
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: VIRAL PROTEIN Ligands: ZN, PO4, A1BXZ |
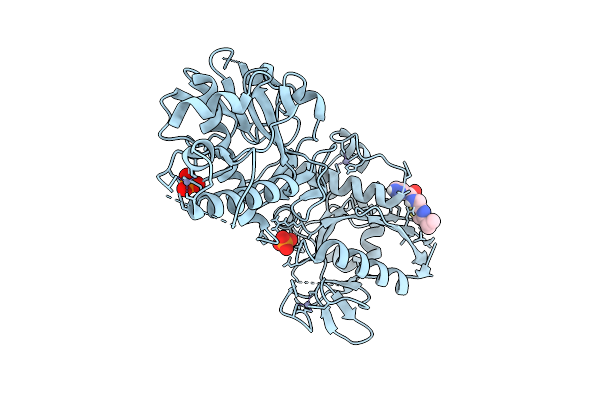 |
Structure Of Sars-Cov-2 Nsp14 Bound To N-((4-Isopropylthiazol-2-Yl)Methyl)-1H-Pyrazole-3-Carboxamide
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: VIRAL PROTEIN Ligands: ZN, PO4, A1BX5 |
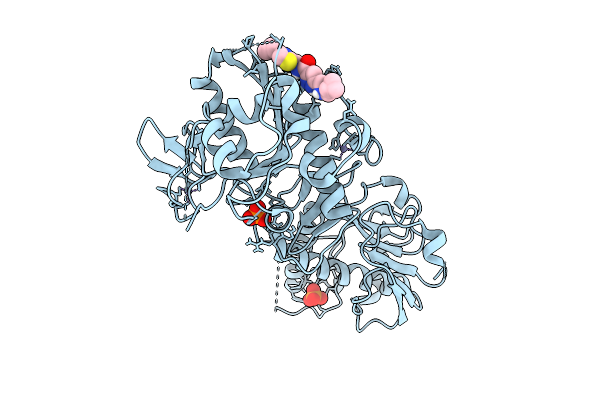 |
Structure Of Sars-Cov-2 Nsp14 Bound To 5-(((Cyclopropylmethyl)Amino)Methyl)-N-((4-Cyclopropylthiazol-2-Yl)Methyl)-1H-Pyrazole-3-Carboxamide
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: VIRAL PROTEIN Ligands: ZN, PO4, A1BYF |
 |
Sars-Cov-2 Nsp14 Bound To N-((2-Ethynylthiazol-4-Yl)Methyl)-1H-Pyrazole-3-Carboxamide
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: VIRAL PROTEIN Ligands: ZN, PO4, A1BYG |
 |
Structure Of Sars-Cov-2 Nsp14 Bound To N-((4-Cyclopropylthiazol-2-Yl)Methyl)-1H-Pyrazolo[3,4-B]Pyridine-3-Carboxamide
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: VIRAL PROTEIN Ligands: ZN, PO4, A1BYT |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: STRUCTURAL PROTEIN/INHIBITOR Ligands: A1BW7 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: STRUCTURAL PROTEIN/INHIBITOR Ligands: A1BW8 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: STRUCTURAL PROTEIN/INHIBITOR Ligands: A1BW9 |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2010-08-04 Classification: HYDROLASE Ligands: OAN, PG4, NA, ACT |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2008-12-16 Classification: HYDROLASE Ligands: ACT, NA, P4G |
 |
Structure Of The Rotor Ring Of F-Type Na+-Atpase From Ilyobacter Tartaricus
Organism: Ilyobacter tartaricus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2005-04-12 Classification: MEMBRANE PROTEIN |
 |
Crystal Structure Of The 92-Residue C-Term. Part Of Tonb With Significant Structural Changes Compared To Shorter Fragments
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.13 Å Release Date: 2004-11-30 Classification: PROTEIN TRANSPORT |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2004-04-06 Classification: TRANSPORT PROTEIN |