Search Count: 18
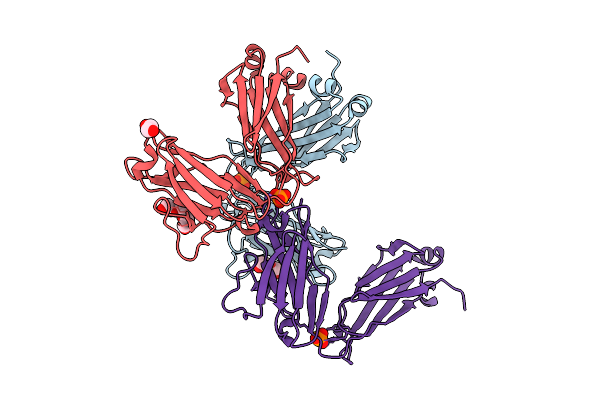 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: IMMUNE SYSTEM Ligands: PEG, PO4, PGE, EDO, NA |
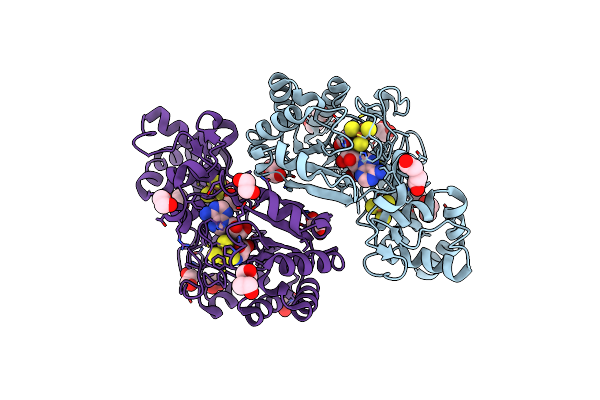 |
Crystal Structure Of Radical Sam Epimerase Epee From Bacillus Subtilis With [4Fe-4S] Clusters And S-Adenosyl-L-Homocysteine Bound.
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Release Date: 2024-01-10 Classification: METAL BINDING PROTEIN Ligands: SF4, SAH, EDO, PEG |
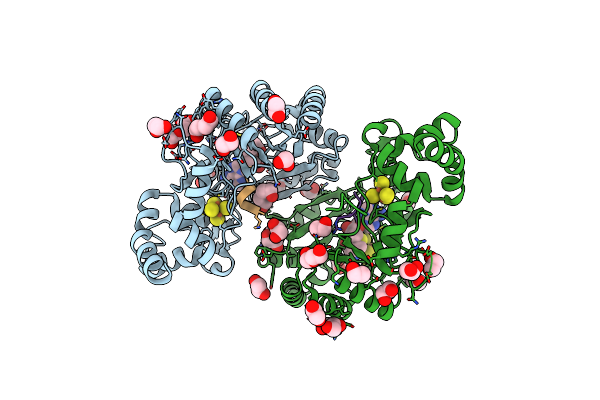 |
Crystal Structure Of Radical Sam Epimerase Epee From Bacillus Subtilis With [4Fe-4S] Clusters, S-Adenosyl-L-Homocysteine And Ripp Peptide 5 Bound
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Release Date: 2024-01-10 Classification: METAL BINDING PROTEIN Ligands: SF4, SAH, EDO, PEG |
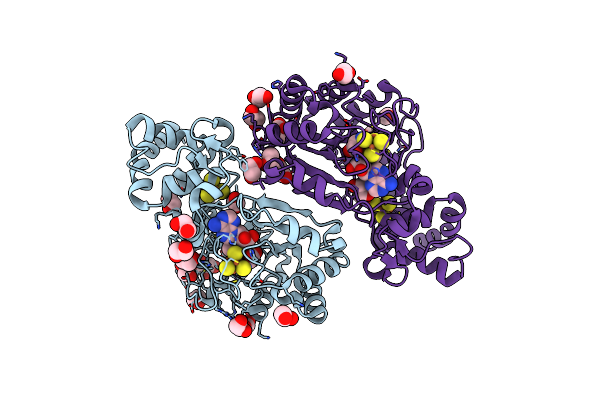 |
Crystal Structure Of Radical Sam Epimerase Epee C223A Mutant From Bacillus Subtilis With [4Fe-4S] Clusters And S-Adenosyl-L-Methionine Bound
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-01-10 Classification: METAL BINDING PROTEIN Ligands: SF4, SAM, EDO, GOL |
 |
Crystal Structure Of Radical Sam Epimerase Epee C223A Mutant From Bacillus Subtilis With [4Fe-4S] Clusters, S-Adenosyl-L-Homocysteine And Ripp Peptide 5 Bound
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Release Date: 2024-01-10 Classification: METAL BINDING PROTEIN Ligands: SF4, SAH, EDO, PEG, GOL |
 |
Crystal Structure Of Radical Sam Epimerase Epee C223A Mutant From Bacillus Subtilis With [4Fe-4S] Clusters, S-Adenosyl-L-Homocysteine And Ripp Peptide 6 Bound
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Release Date: 2024-01-10 Classification: METAL BINDING PROTEIN Ligands: SF4, SAH, EDO, PEG |
 |
Crystal Structure Of Radical Sam Epimerase Epee D210A Mutant From Bacillus Subtilis With [4Fe-4S] Clusters, S-Adenosyl-L-Homocysteine And Persulfurated Cysteine Bound
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-01-10 Classification: METAL BINDING PROTEIN Ligands: SF4, SAH, EDO, PEG |
 |
The Crystal Structure Of Erwinia Tasmaniensis Levansucrase In Complex With (S)-1,2,4-Butanentriol
Organism: Erwinia tasmaniensis (strain dsm 17950 / cip 109463 / et1/99)
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2022-06-22 Classification: TRANSFERASE Ligands: ZN, 0V1 |
 |
Crystal Structure Of Malus Domestica Double Bond Reductase (Mddbr) Apo Form
Organism: Pyrus ussuriensis x pyrus communis
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2021-02-03 Classification: BIOSYNTHETIC PROTEIN Ligands: SO4 |
 |
Crystal Structure Of Malus Domestica Double Bond Reductase (Mddbr) In Complex With Nadph
Organism: Malus domestica
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2021-02-03 Classification: BIOSYNTHETIC PROTEIN Ligands: NAP, MPD, EDO, PGR, NA |
 |
Crystal Structure Of Malus Domestica Double Bond Reductase (Mddbr) Ternary Complex
Organism: Malus domestica
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2021-02-03 Classification: BIOSYNTHETIC PROTEIN Ligands: NAP, MPD, EDO, FER, NA |
 |
X-Ray Structure Of The Levansucrase From Erwinia Tasmaniensis In Complex With Levanbiose
Organism: Erwinia tasmaniensis (strain dsm 17950 / cip 109463 / et1/99)
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2020-04-01 Classification: TRANSFERASE Ligands: GOL, ZN |
 |
Organism: Erwinia tasmaniensis
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2019-02-06 Classification: TRANSFERASE Ligands: GOL, ZN |
 |
The Crystal Structure Of Dfoa Bound To Fad, The Desferrioxamine Biosynthetic Pathway Cadaverine Monooxygenase From The Fire Blight Disease Pathogen Erwinia Amylovora
Organism: Erwinia amylovora cfbp1430
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2018-06-27 Classification: BIOSYNTHETIC PROTEIN Ligands: FAD |
 |
The Crystal Structure Of Dfoj, The Desferrioxamine Biosynthetic Pathway Lysine Decarboxylase From The Fire Blight Disease Pathogen Erwinia Amylovora
Organism: Erwinia amylovora (strain cfbp1430)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-02-28 Classification: LYASE Ligands: PLP |
 |
The Crystal Structure Of Dfoc, The Desferrioxamine Biosynthetic Pathway Acetyltransferase/Non-Ribosomal Peptide Synthetase (Nrps)-Independent Siderophore (Nis) From The Fire Blight Disease Pathogen Erwinia Amylovora
Organism: Erwinia amylovora cfbp1430
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2018-02-28 Classification: BIOSYNTHETIC PROTEIN |
 |
The Crystal Structure Of Dfoa Bound To Fad And Nadp; The Desferrioxamine Biosynthetic Pathway Cadaverine Monooxygenase From The Fire Blight Disease Pathogen Erwinia Amylovora
Organism: Erwinia amylovora cfbp1430
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2018-02-28 Classification: BIOSYNTHETIC PROTEIN Ligands: FAD, NAP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2016-01-27 Classification: TRANSPORT PROTEIN Ligands: IPJ, EDO |

