Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The Ground State
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLC, LFA, GOL, RET, PO4 |
 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The P596 State
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLC, LFA, GOL, RET, PO4 |
 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) - Human Gtpase Arf1 (L8K,Q71L) Chimera; Ground State
Organism: Candidatus actinomarina minuta, Homo sapiens, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: GDP, LFA, RET |
 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) - Human Gtpase Arf1 (L8K,Q71L) Chimera; N State
Organism: Candidatus actinomarina minuta, Homo sapiens, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: GDP, LFA, OLA, RET |
 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The O* State
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLA, LFA, RET |
 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The O* State, Ph 8.8
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLA, LFA, RET |
 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The O State Obtained By Cryotrapping
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLA, LFA, RET |
 |
Structure Of E. Coli Glpg In Complex With Peptide Derived Inhibitor Ac-Vrha-Conh-[4-(4-Butyl)-Phenoxy-1-Phenyl-2-Butyl]
Organism: Escherichia coli k-12, Providencia stuartii
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2023-10-25 Classification: HYDROLASE |
 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The M-Like State
Organism: Candidatus actinomarina minuta
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-06-01 Classification: MEMBRANE PROTEIN Ligands: LFA, OLB, OLC, RET |
 |
Structure Of Marine Actinobacteria Clade Rhodopsin (Macr) In Orange Form In P1211 Space Group
Organism: Candidatus actinomarina minuta
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-06-01 Classification: MEMBRANE PROTEIN Ligands: LFA, OLB, OLC |
 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The O State
Organism: Candidatus actinomarina minuta
Method: X-RAY DIFFRACTION Resolution:1.09 Å Release Date: 2022-06-01 Classification: MEMBRANE PROTEIN Ligands: LFA, RET |
 |
Crystal Structure Of A Light-Driven Proton Pump Lr (Mac) From Leptosphaeria Maculans
Organism: Leptosphaeria maculans
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2021-07-07 Classification: MEMBRANE PROTEIN Ligands: LFA, OLA |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2020-05-06 Classification: MEMBRANE PROTEIN |
 |
Crystal Structure Of A Proteolytic Fragment Of Narq Comprising Sensor And Tm Domains
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-03-04 Classification: TRANSFERASE Ligands: NO3 |
 |
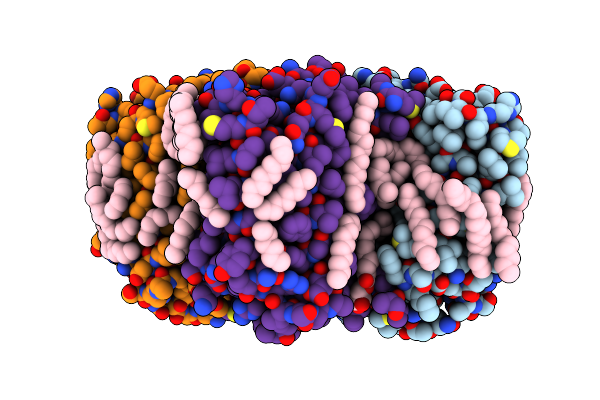
Crystal Structure Of The C14 Ring Of The F1Fo Atp Synthase From Spinach Chloroplast
Organism: Spinacia oleracea
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-12-25 Classification: MEMBRANE PROTEIN Ligands: LFA, OLC |
 |
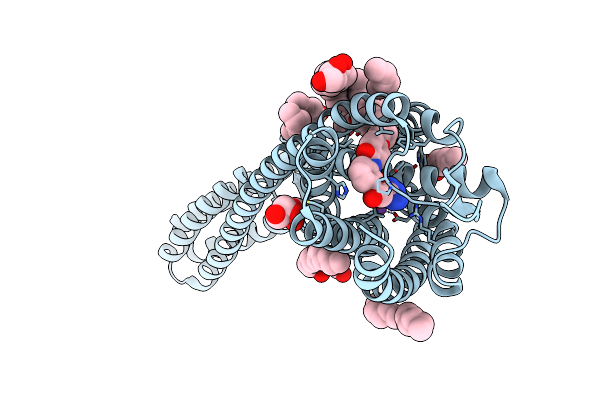
Organism: Organic lake phycodnavirus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-11-06 Classification: MEMBRANE PROTEIN Ligands: LFA, RET, OLB |
 |
Crystal Structure Of Cysteinyl Leukotriene Receptor 1 In Complex With Pranlukast
Organism: Homo sapiens, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2019-10-30 Classification: MEMBRANE PROTEIN Ligands: KNT, NA, OLC, OLA |
 |
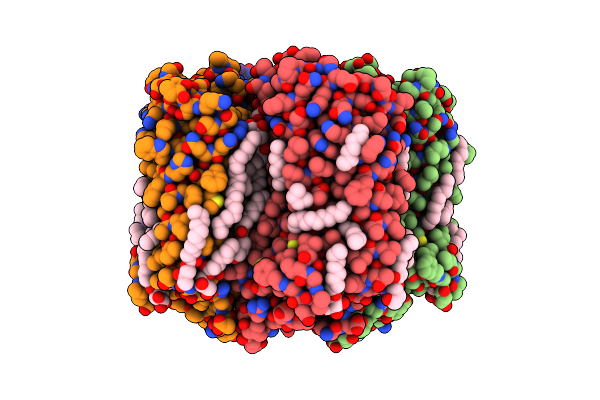
Xfel Crystal Structure Of The Human Cysteinyl Leukotriene Receptor 1 In Complex With Zafirlukast
Organism: Homo sapiens, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.53 Å Release Date: 2019-10-30 Classification: MEMBRANE PROTEIN Ligands: ZLK, OLC, OLA, 1PE, NA |
 |
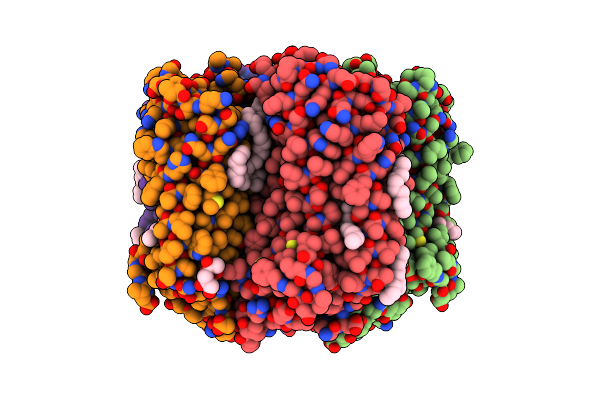
Crystal Structure Of The Light-Driven Sodium Pump Kr2 In The Pentameric Form, Ph 6.0
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2019-04-24 Classification: MEMBRANE PROTEIN Ligands: OLC, LFA, NA, BOG, RET |
 |
Crystal Structure Of The Light-Driven Sodium Pump Kr2 In The Pentameric Form, Ph 5.0
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2019-04-24 Classification: MEMBRANE PROTEIN Ligands: LFA, OLC, NA, RET |