Search Count: 21
 |
Structure Of Hyoscyamine 6-Beta Hydroxylase In Complex With Iron, 2-Oxoglutarate, And Hyoscyamine
Organism: Atropa belladonna
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2023-11-22 Classification: OXIDOREDUCTASE Ligands: SR, EDO, FMT, FE2, AKG, HYO |
 |
Structure Of Hyoscyamine 6-Beta Hydroxylase In Complex With Vanadyl, Succinate, And Hyoscyamine
Organism: Atropa belladonna
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2023-11-22 Classification: OXIDOREDUCTASE Ligands: EDO, HYO, SIN, FMT, V, O, SR |
 |
Structure Of Hyoscyamine 6-Beta Hydroxylase In Complex With Iron, Succinate, And 6-Oh-Hyoscyamine
Organism: Atropa belladonna
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2023-11-22 Classification: OXIDOREDUCTASE Ligands: FE2, EDO, FMT, OVR, SIN, SR |
 |
Structure Of Hyoscyamine 6-Beta Hydroxylase In Complex With Iron, 2-Oxoglutarate, And 6-Oh-Hyoscyamine
Organism: Atropa belladonna
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2023-11-22 Classification: OXIDOREDUCTASE Ligands: FE2, EDO, FMT, AKG, OVR, SR |
 |
Structure Of Hyoscyamine 6-Beta Hydroxylase In Complex With Vanadyl, Succinate, And 6-Oh-Hyoscyamine
Organism: Atropa belladonna
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2023-11-22 Classification: OXIDOREDUCTASE Ligands: EDO, SIN, FMT, OVR, V, O, SR |
 |
Structure Of Hyoscyamine 6-Beta Hydroxylase In Complex With Iron, Succinate, And Scopolamine
Organism: Atropa belladonna
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2023-11-22 Classification: OXIDOREDUCTASE Ligands: FE2, EDO, FMT, OW0, SIN, SR |
 |
Structure Of L289F Hyoscyamine 6-Beta Hydroxylase In Complex With Iron, 2-Oxoglutarate, And Hyoscyamine
Organism: Atropa belladonna
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2023-11-22 Classification: OXIDOREDUCTASE Ligands: FE2, EDO, FMT, HYO, AKG, SR |
 |
Structure Of L289F Hyoscyamine 6-Beta Hydroxylase In Complex With Vanadyl, Succinate, And Hyoscyamine
Organism: Atropa belladonna
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2023-11-22 Classification: OXIDOREDUCTASE Ligands: EDO, FMT, HYO, SIN, V, O, SR |
 |
Structure Of L289F Hyoscyamine 6-Beta Hydroxylase In Complex With Iron, 2-Oxoglutarate, And 6-Oh-Hyoscyamine
Organism: Atropa belladonna
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2023-11-22 Classification: OXIDOREDUCTASE Ligands: FE2, EDO, FMT, AKG, OVR, SR |
 |
Structure Of L289F Hyoscyamine 6-Beta Hydroxylase In Complex With Vanadyl, Succinate, And 6-Oh-Hyoscyamine
Organism: Atropa belladonna
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2023-11-22 Classification: OXIDOREDUCTASE Ligands: EDO, FMT, SIN, OVR, V, SR |
 |
X-Ray Crystal Structure Of Flavobacterium Johnsoniae Dimanganese(Ii) Ribonucleotide Reductase Beta Subunit (Aerobic)
Organism: Flavobacterium johnsoniae (strain atcc 17061 / dsm 2064 / uw101)
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2018-06-27 Classification: OXIDOREDUCTASE Ligands: MN, MG |
 |
X-Ray Crystal Structure Of Flavobacterium Johnsoniae Dimanganese(Ii) Ribonucleotide Reductase Beta Subunit (Anaerobic)
Organism: Flavobacterium johnsoniae (strain atcc 17061 / dsm 2064 / uw101), Flavobacterium johnsoniae
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2018-04-18 Classification: OXIDOREDUCTASE Ligands: MN |
 |
X-Ray Crystal Structure Of Flavobacterium Johnsoniae Dimanganese(Ii) Ribonucleotide Reductase Beta Subunit (As-Isolated)
Organism: Flavobacterium johnsoniae (strain atcc 17061 / dsm 2064 / uw101)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-04-18 Classification: OXIDOREDUCTASE Ligands: MN, MG |
 |
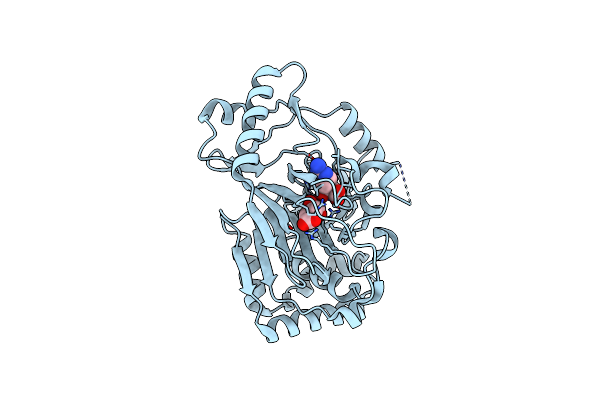
Vioc L-Arginine Hydroxylase Bound To Fe(Ii), L-Arginine, And 2-Oxo-Glutaric Acid
Organism: Streptomyces vinaceus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2017-09-06 Classification: OXIDOREDUCTASE Ligands: FE2, AKG, ARG |
 |
Vioc L-Arginine Hydroxylase Bound To Fe(Ii), 3S-Hydroxy-L-Arginine, And 2Og
Organism: Streptomyces vinaceus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-09-06 Classification: OXIDOREDUCTASE Ligands: AKG, FE2, ZZU |
 |
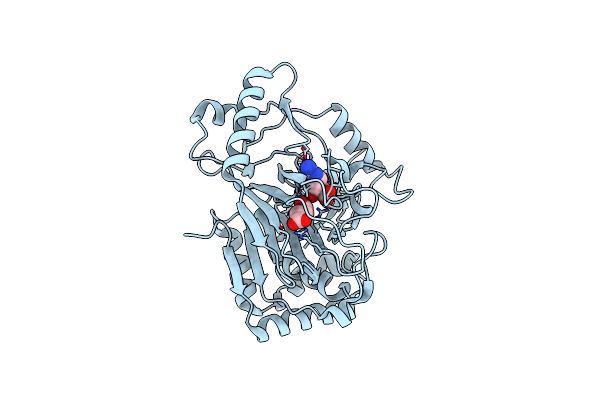
Vioc L-Arginine Hydroxylase Bound To Fe(Ii), L-Arginine, And A Peroxysuccinate Intermediate
Organism: Streptomyces vinaceus
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2017-09-06 Classification: OXIDOREDUCTASE Ligands: FE2, ARG, OKG |
 |
Vioc L-Arginine Hydroxylase Bound To Fe(Ii), 3S-Hydroxy-L-Arginine, And Succinate
Organism: Streptomyces vinaceus
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2017-09-06 Classification: OXIDOREDUCTASE Ligands: FE2, SIN, ZZU |
 |
Organism: Streptomyces vinaceus
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2017-09-06 Classification: OXIDOREDUCTASE Ligands: FE2, SIN, ARG |
 |
Vioc L-Arginine Hydroxylase Bound To The Vanadyl Ion, L-Arginine, And Succinate
Organism: Streptomyces vinaceus
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2017-09-06 Classification: OXIDOREDUCTASE Ligands: SIN, ARG, VVO |
 |
Structure Of Pas-Gaf Fragment Of Deinococcus Phytochrome By Serial Femtosecond Crystallography
Organism: Deinococcus radiodurans (strain atcc 13939 / dsm 20539 / jcm 16871 / lmg 4051 / nbrc 15346 / ncimb 9279 / r1 / vkm b-1422)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2017-02-22 Classification: TRANSFERASE Ligands: LBV, NI, CL, EDO |

