Search Count: 18
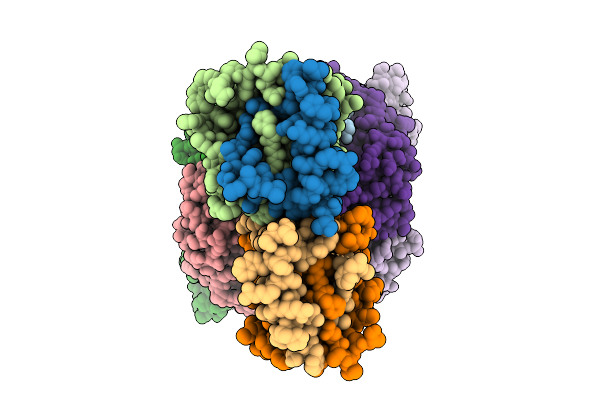 |
Thermus Thermophilus Mrec-Mred Complex With An Internal Mred Bril Fusion And An Anti-Bril Fab
Organism: Escherichia coli, Thermus thermophilus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: MEMBRANE PROTEIN |
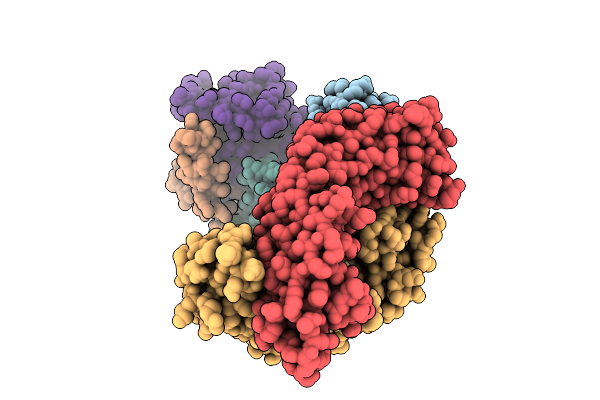 |
Thermus Thermophilus Mrec-Mred Complex With A C-Terminal Mred Bril Fusion And An Anti-Bril Fab
Organism: Escherichia coli, Thermus thermophilus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: MEMBRANE PROTEIN |
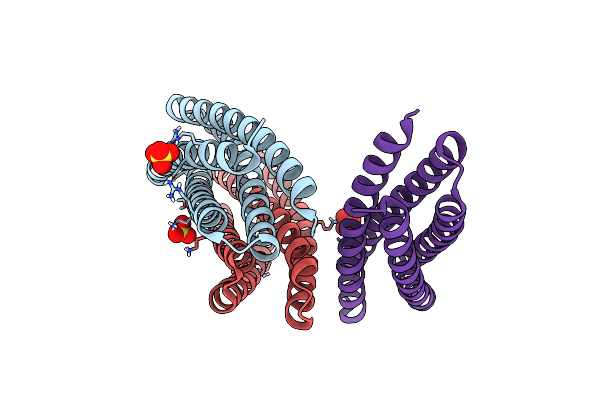 |
De Novo Designed Protein Binds Poly Adp Ribose Polymerase Inhibitors (Parpi) - Apo
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2024-04-24 Classification: DE NOVO PROTEIN Ligands: SO4 |
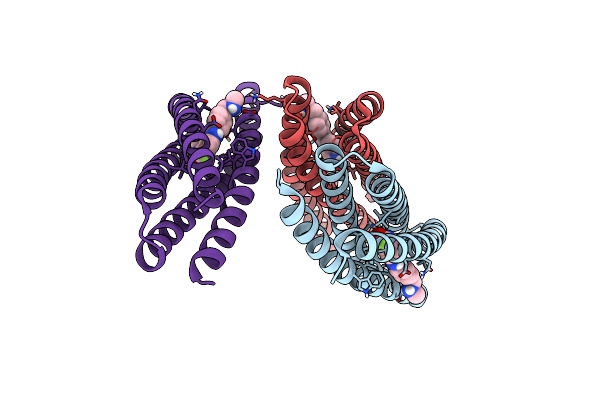 |
De Novo Designed Protein Binds Poly Adp Ribose Polymerase Inhibitors (Parpi) - Holo Rucaparib
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2024-04-24 Classification: DE NOVO PROTEIN Ligands: RPB |
 |
De Novo Designed Protein Binds Poly Adp Ribose Polymerase Inhibitors (Parpi) - Holo Mefuparib
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2024-04-24 Classification: DE NOVO PROTEIN Ligands: IQM |
 |
De Novo Designed Protein Binds Poly Adp Ribose Polymerase Inhibitors (Parpi) - Holo Niraparib
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2024-04-24 Classification: DE NOVO PROTEIN Ligands: 3JD |
 |
De Novo Designed Protein Binds Poly Adp Ribose Polymerase Inhibitors (Parpi) - Holo Veliparib
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.29 Å Release Date: 2024-04-24 Classification: DE NOVO PROTEIN Ligands: 78P, SO4 |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2020-12-09 Classification: DE NOVO PROTEIN Ligands: ZN, 7BU, 2PE |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2020-08-26 Classification: DE NOVO PROTEIN Ligands: ACT, SO4 |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2020-08-26 Classification: DE NOVO PROTEIN Ligands: SO4, ACT, GG2 |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-08-26 Classification: DE NOVO PROTEIN |
 |
Organism: Influenza a virus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-09-19 Classification: MEMBRANE PROTEIN Ligands: 308, CL |
 |
Organism: Influenza a virus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-09-19 Classification: MEMBRANE PROTEIN Ligands: EU7, RIM |
 |
Organism: Influenza a virus
Method: X-RAY DIFFRACTION Resolution:2.63 Å Release Date: 2018-09-19 Classification: MEMBRANE PROTEIN Ligands: E01, CL |
 |
Influenza A M2 Transmembrane Domain Bound To Rimantadine In The Inward(Open) Conformation
Organism: Influenza a virus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2018-09-19 Classification: MEMBRANE PROTEIN Ligands: CL, EU7, RIM, NA |
 |
Organism: Escherichia coli
Method: SOLUTION NMR Release Date: 2017-08-09 Classification: UNKNOWN FUNCTION |
 |
Organism: Escherichia coli
Method: SOLUTION NMR Release Date: 2017-08-09 Classification: UNKNOWN FUNCTION Ligands: 7BU |
 |
Zinc-Binding Structure Of A Catalytic Amyloid From Solid-State Nmr Spectroscopy
Organism: Synthetic construct
Method: SOLID-STATE NMR Release Date: 2017-05-31 Classification: METAL BINDING PROTEIN Ligands: ZN |

