Search Count: 349
 |
Organism: Polaromonas sp. (strain js666 / atcc baa-500)
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2021-04-07 Classification: OXIDOREDUCTASE Ligands: HEC, EDO |
 |
Organism: Polaromonas sp.
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2021-04-07 Classification: OXIDOREDUCTASE Ligands: HEM, OC9 |
 |
Organism: Polaromonas sp. js666
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2015-01-21 Classification: OXIDOREDUCTASE Ligands: CL, NA |
 |
Crystal Structure Of A Tctc Solute Binding Protein From Polaromonas (Bpro_3516, Target Efi-510338), No Ligand
Organism: Polaromonas sp. (strain js666 / atcc baa-500)
Method: X-RAY DIFFRACTION Resolution:1.24 Å Release Date: 2014-12-24 Classification: UNKNOWN FUNCTION Ligands: CL |
 |
Crystal Structure Of A Trap Periplasmic Solute Binding Protein From Polaromonas Sp Js666 (Bpro_0088, Target Efi-510167) Bound To D-Erythronate
Organism: Polaromonas sp. js666
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2014-05-14 Classification: SOLUTE-BINDING PROTEIN Ligands: EAX, MLA, FMT |
 |
Crystal Structure Of A Trap Periplasmic Solute Binding Protein From Polaromonas Sp Js666 (Bpro_1871, Target Efi-510164) Bound To D-Erythronate
Organism: Polaromonas sp. js666
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-05-14 Classification: SOLUTE-BINDING PROTEIN Ligands: EAX |
 |
Crystal Structure Of A Trap Periplasmic Solute Binding Protein From Polaromonas Sp. Js666 (Bpro_4736), Target Efi-510156, With Bound Benzoyl Formate, Space Group P21
Organism: Polaromonas sp.
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2013-09-25 Classification: TRANSPORT PROTEIN Ligands: 173, SO4 |
 |
Crystal Structure Of A Trap Periplasmic Solute Binding Protein From Polaromonas Sp. Js666 (Bpro_4736), Target Efi-510156, With Bound Benzoyl Formate, Space Group P6522
Organism: Polaromonas sp.
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-09-25 Classification: TRANSPORT PROTEIN Ligands: 173 |
 |
Organism: Polaromonas sp. js666
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2013-05-01 Classification: LYASE Ligands: GOL |
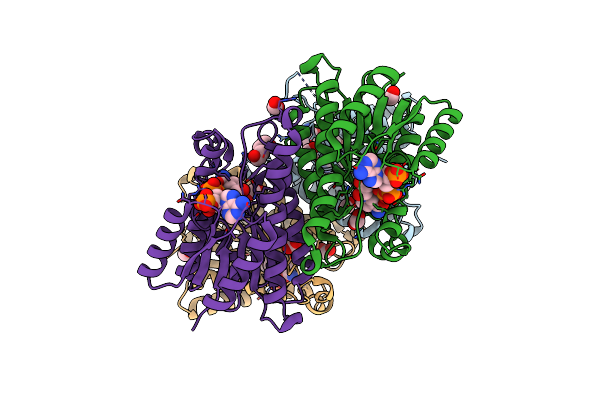 |
Crystal Structure Of Bpro0239 Oxidoreductase From Polaromonas Sp. Js666 In Nadp Bound Form
Organism: Polaromonas sp. js666
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2013-01-30 Classification: OXIDOREDUCTASE Ligands: FMT, NAP, GOL, PEG |
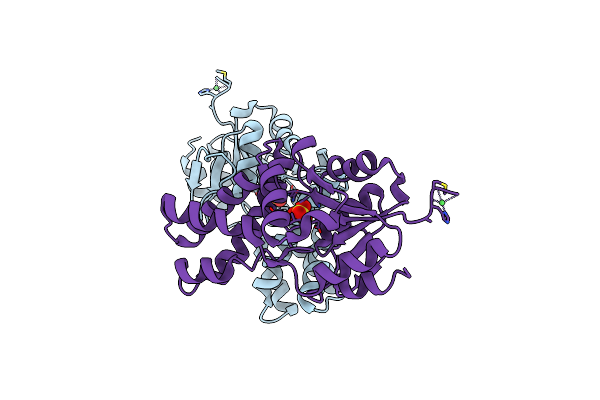 |
Organism: Polaromonas sp. js666
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2012-11-14 Classification: HYDROLASE Ligands: NI, SO4 |
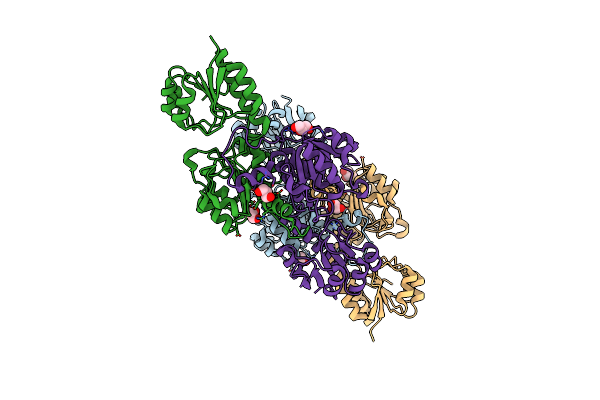 |
Crystal Structure Of Putative D-Isomer Specific 2-Hydroxyacid Dehydrogenase, Nad-Binding From Polaromonas Sp. Js6 66
Organism: Polaromonas sp. js666
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2012-07-25 Classification: OXIDOREDUCTASE Ligands: GOL, CL |
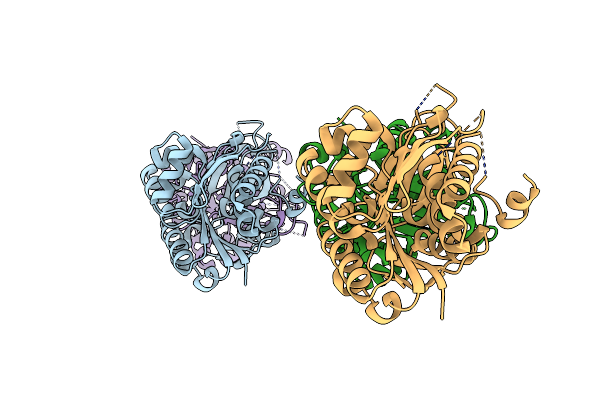 |
Organism: Polaromonas sp. js666
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2012-03-21 Classification: TRANSFERASE Ligands: CL |
 |
Crystal Structure Of A 2-Hydroxy-3-Oxopropionate Reductase From Polaromonas Sp. Js666
Organism: Polaromonas sp. js666
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2012-02-15 Classification: OXIDOREDUCTASE Ligands: SO4 |
 |
Crystal Structure Of Putative Amidohydrolase-2 (Efi-Target 500288)From Polaromonas Sp. Js666
Organism: Polaromonas sp. js666
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-08-24 Classification: HYDROLASE Ligands: EPE, SO4, ACT, CL, GOL |
 |
Organism: Polaromonas sp. js666
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2009-04-14 Classification: TRANSFERASE |
 |
Crystal Structure Of L-Talarate Dehydratase From Polaromonas Sp. Js666 Complexed With Mg And L-Glucarate
Organism: Polaromonas sp.
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2008-03-04 Classification: ISOMERASE Ligands: MG, LGT |
 |
Crystal Structure Of A Member Of Enolase Superfamily From Polaromonas Sp. Js666
Organism: Polaromonas sp.
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2007-12-18 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION Ligands: MG |
 |
Crystal Structure Of Mandelate Racemase/Muconate Lactonizing Enzyme From Polaromonas Sp. Js666
Organism: Polaromonas sp.
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2007-02-27 Classification: ISOMERASE Ligands: CA |
 |

