Search Count: 153
 |
Organism: Podospora anserina (strain s / atcc mya-4624 / dsm 980 / fgsc 10383)
Method: SOLID-STATE NMR Release Date: 2018-10-10 Classification: PROTEIN FIBRIL |
 |
Organism: Podospora anserina
Method: SOLUTION NMR Release Date: 2017-02-01 Classification: PROTEIN FIBRIL Ligands: 3LS |
 |
Crystal Structure Of Laglidadg Meganuclease I-Panmi With Coordinated Calcium Ions
Organism: Podospora anserina, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2016-03-30 Classification: HYDROLASE/DNA Ligands: CA, GOL |
 |
Q17M Crystal Structure Of Podosopora Anserina Putative Kinesin Light Chain Nearly Identical Tpr-Like Repeats
Organism: Podospora anserina
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2015-10-07 Classification: UNKNOWN FUNCTION Ligands: SO4 |
 |
Crystal Structure Of Podosopora Anserina Putative Kinesin Light Chain Nearly Identical Tpr-Like Repeats
Organism: Podospora anserina
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2015-10-07 Classification: UNKNOWN FUNCTION |
 |
Organism: Podospora anserina
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2015-05-27 Classification: TRANSFERASE Ligands: EDO |
 |
Organism: Podospora anserina
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2015-05-27 Classification: TRANSFERASE Ligands: SAM, PO4, PG4, MG |
 |
Organism: Podospora anserina
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2015-05-27 Classification: TRANSFERASE Ligands: SAH, PEG |
 |
Crystal Structure Of The Alpha-L-Arabinofuranosidase Paabf62A From Podospora Anserina In Complex With Cellotriose
Organism: Podospora anserina
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-01-15 Classification: HYDROLASE Ligands: CA, TRS, 1PE, EPE |
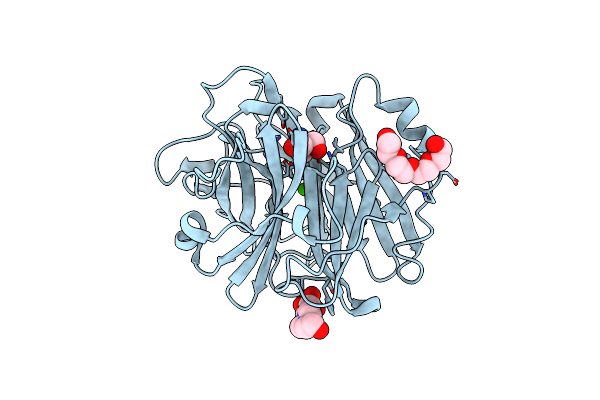 |
Crystal Structure Of The Alpha-L-Arabinofuranosidase Paabf62A From Podospora Anserina
Organism: Podospora anserina
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2014-01-15 Classification: HYDROLASE Ligands: CA, EPE, TRS, 1PE |
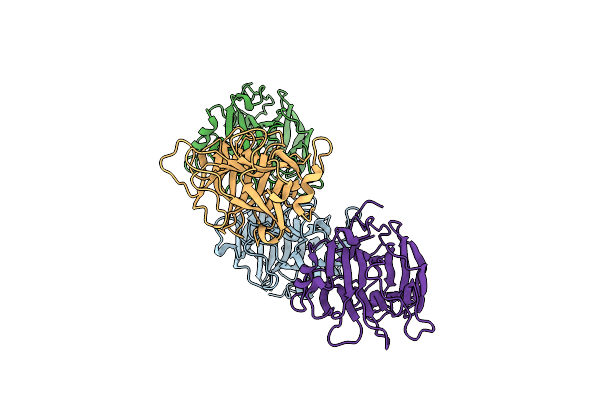 |
Crystal Structure Of A Gh131 Beta-Glucanase Catalytic Domain From Podospora Anserina
Organism: Podospora anserina
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2013-09-11 Classification: HYDROLASE |
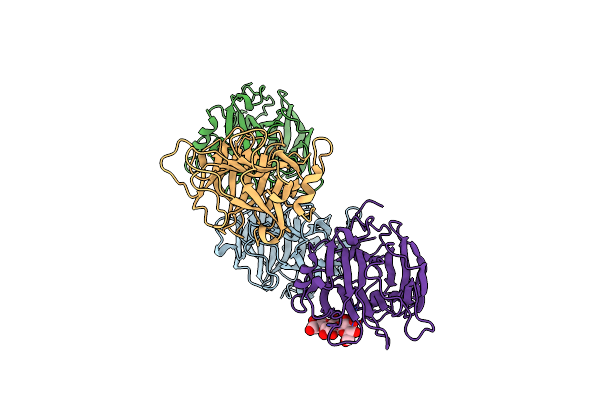 |
Organism: Podospora anserina
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2013-09-11 Classification: HYDROLASE |
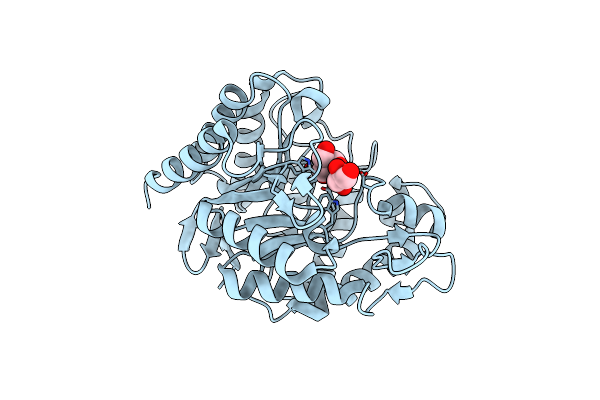 |
Organism: Podospora anserina
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2013-04-17 Classification: HYDROLASE Ligands: GOL, TRS |
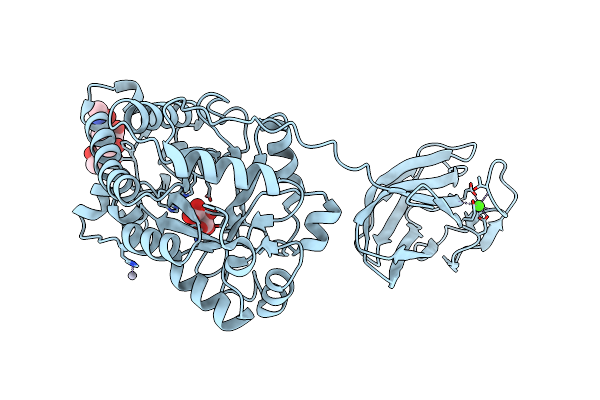 |
Organism: Podospora anserina
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2013-04-17 Classification: HYDROLASE Ligands: CA, HG, TLA |
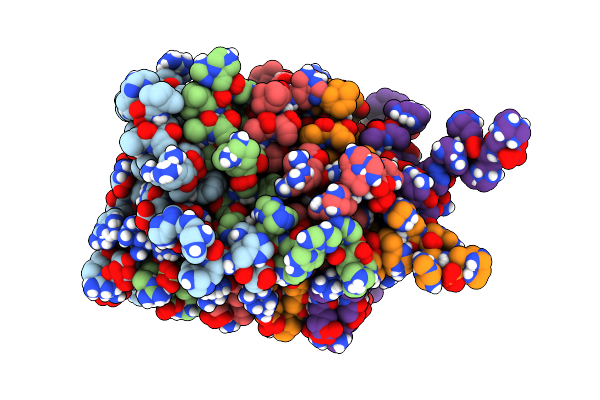 |
Organism: Podospora anserina
Method: SOLID-STATE NMR Release Date: 2011-06-01 Classification: PROTEIN FIBRIL Ligands: CGO |
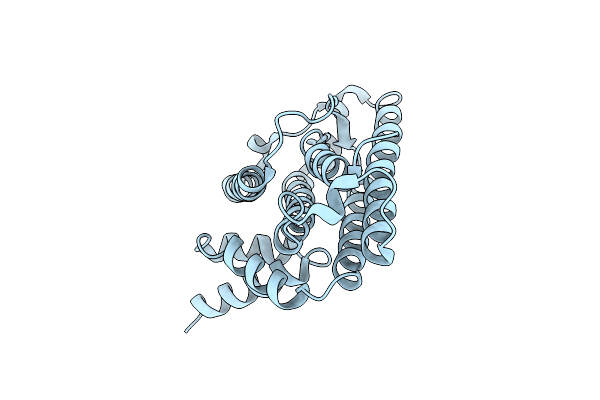 |
Organism: Podospora anserina
Method: X-RAY DIFFRACTION Resolution:2.62 Å Release Date: 2010-07-28 Classification: PRION-BINDING PROTEIN |
 |
Organism: Podospora anserina
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2010-07-28 Classification: PRION-BINDING PROTEIN Ligands: CL |
 |
Organism: Podospora anserina
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2010-07-28 Classification: PRION-BINDING PROTEIN Ligands: DTT, DTU |
 |
High-Resolution Structure Of The Het-S(218-289) Prion In Its Amyloid Form Obtained By Solid-State Nmr
Organism: Podospora anserina
Method: SOLID-STATE NMR Release Date: 2010-06-02 Classification: PROTEIN FIBRIL |
 |
Organism: Podospora anserina
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2010-06-02 Classification: OXIDOREDUCTASE Ligands: FAD, MG, SO4 |

