Search Count: 121
 |
The Structure Of The Glycopeptidase Catalytic Domain Including The Linker Of Amuc_1438
Organism: Akkermansia muciniphila
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-08-31 Classification: HYDROLASE Ligands: ZN, NA, EDO |
 |
Organism: Akkermansia muciniphila
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2022-08-31 Classification: HYDROLASE Ligands: ZN, CA, NA |
 |
Organism: Bacteroides plebeius
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2022-02-09 Classification: OXIDOREDUCTASE |
 |
Structure Of The Exo-L-Galactose-6-Sulfatase Bus1_11 From Bacteroides Uniformis
Organism: Bacteroides uniformis
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-02-09 Classification: HYDROLASE Ligands: CA, NI |
 |
Structure Of Exo-L-Galactose-6-Sulfatase Bus1_11 From Bacteroides Uniformis In Complex With Neoporphyrabiose
Organism: Bacteroides uniformis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2022-02-09 Classification: HYDROLASE Ligands: CA, NA |
 |
Structure Of The Exo-Alpha-L-Galactosidase Bpgh29 From Bacteroides Plebeius
Organism: Bacteroides plebeius
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-02-09 Classification: HYDROLASE Ligands: EDO, CA |
 |
Structure Of The Exo-Alpha-L-Galactosidase Bpgh29 From Bacteroides Plebeius In Complex With L-Galactose
Organism: Bacteroides plebeius
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2022-02-09 Classification: HYDROLASE Ligands: CA, GXL, EDO |
 |
Structure Of The Exo-Alpha-L-Galactosidase Bpgh29 (D264N Mutant) From Bacteroides Plebeius In Complex With Paranitrophenyl-Alpha-L-Galactopyranoside
Organism: Bacteroides plebeius
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2022-02-09 Classification: HYDROLASE Ligands: CA, Y9Y, NI, NA, CL |
 |
The Structure Of The Catalytic Module Of The Metalloprotease Zmpa From Clostridium Perfringens
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-02-03 Classification: HYDROLASE Ligands: MES, EDO |
 |
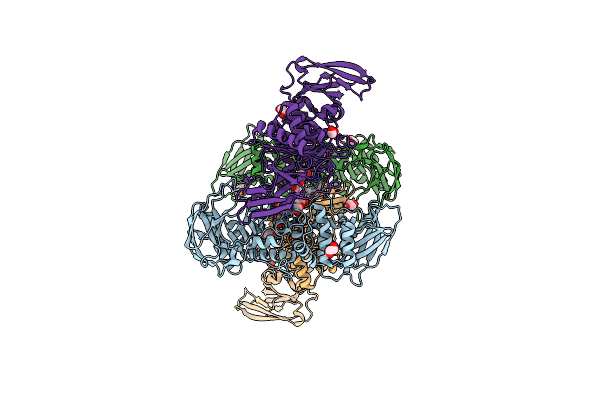
The Structure Of The M60 Catalytic Domain From Clostridium Perfringens Zmpc
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2021-02-03 Classification: HYDROLASE Ligands: ZN, EDO |
 |
The Structure Of The M60 Catalytic Domain From Clostridium Perfringens Zmpc In Complex The Sialyl T Antigen
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2021-02-03 Classification: HYDROLASE Ligands: ZN, EDO, SER |
 |
The Structure Of The Cbm32-1, Cbm32-2, And M60 Catalytic Domains From Clostridium Perfringens Zmpb
Organism: Clostridium perfringens (strain atcc 13124 / dsm 756 / jcm 1290 / ncimb 6125 / nctc 8237 / type a)
Method: X-RAY DIFFRACTION Resolution:4.60 Å Release Date: 2021-02-03 Classification: HYDROLASE Ligands: ZN |
 |
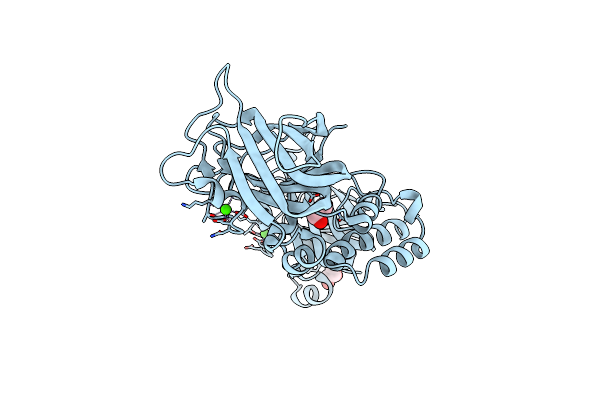
The Structure Of Cbm32-1 And Cbm32-2 Domains From Clostridium Perfringens Zmpb In Complex With Galnac
Organism: Clostridium perfringens (strain atcc 13124 / dsm 756 / jcm 1290 / ncimb 6125 / nctc 8237 / type a)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-02-03 Classification: SUGAR BINDING PROTEIN Ligands: CA, A2G, GOL |
 |
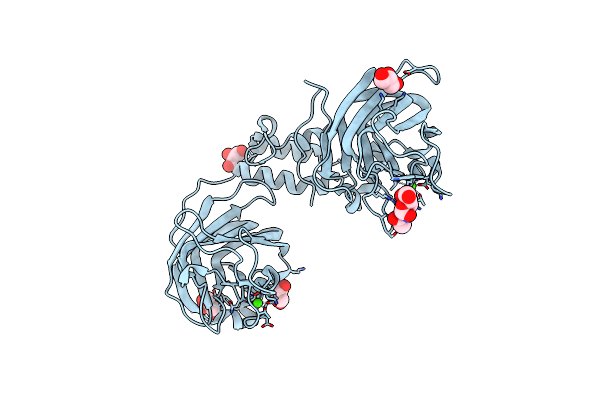
The Structure Of Cbm32-1 And Cbm32-2 Domains From Clostridium Perfringens Zmpb
Organism: Clostridium perfringens (strain atcc 13124 / dsm 756 / jcm 1290 / ncimb 6125 / nctc 8237 / type a)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-02-03 Classification: SUGAR BINDING PROTEIN Ligands: CA, GOL |
 |
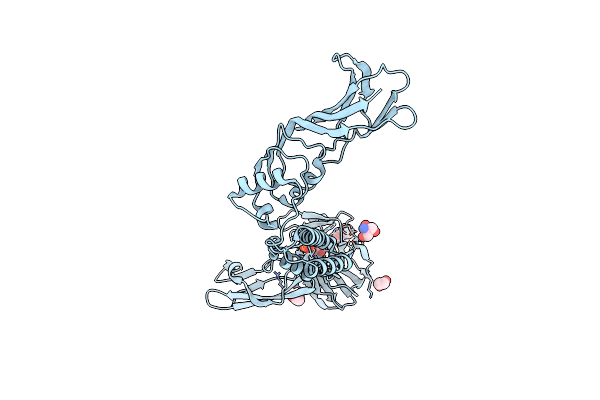
The Structure Of Cbm32-1 And Cbm32-2 Domains From Clostridium Perfringens Zmpb In Complex With Galnac
Organism: Clostridium perfringens (strain atcc 13124 / dsm 756 / jcm 1290 / ncimb 6125 / nctc 8237 / type a)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-02-03 Classification: SUGAR BINDING PROTEIN Ligands: A2G, CA, GOL |
 |
The Structure Of Cbm51-2 In Complex With Glcnac And Int Domains From Clostridium Perfringens Zmpb
Organism: Clostridium perfringens (strain atcc 13124 / dsm 756 / jcm 1290 / ncimb 6125 / nctc 8237 / type a)
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2021-02-03 Classification: SUGAR BINDING PROTEIN Ligands: CA, NAG, EDO, PO4, TRS |
 |
Organism: Clostridium perfringens (strain atcc 13124 / dsm 756 / jcm 1290 / ncimb 6125 / nctc 8237 / type a)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-02-03 Classification: SUGAR BINDING PROTEIN Ligands: CA |
 |
The Structure Of The M60 Catalytic Domain With The Cbm51-1 And Cbm51-2 Domains From Clostridium Perfringens Zmpb
Organism: Clostridium perfringens (strain atcc 13124 / dsm 756 / jcm 1290 / ncimb 6125 / nctc 8237 / type a)
Method: X-RAY DIFFRACTION Resolution:4.60 Å Release Date: 2021-02-03 Classification: HYDROLASE |
 |
Organism: Pseudoalteromonas fuliginea
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-09-16 Classification: HYDROLASE Ligands: EDO |
 |
Organism: Streptococcus pneumoniae serotype 4 (strain atcc baa-334 / tigr4)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2019-10-02 Classification: HYDROLASE Ligands: EDO, TLA |

