Search Count: 22
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: ONCOPROTEIN Ligands: ZN, A1IWG |
 |
Organism: Danio rerio
Method: X-RAY DIFFRACTION Resolution:0.98 Å Release Date: 2022-11-16 Classification: DNA BINDING PROTEIN Ligands: K |
 |
Organism: Danio rerio
Method: X-RAY DIFFRACTION Resolution:1.18 Å Release Date: 2022-11-16 Classification: DNA BINDING PROTEIN Ligands: K |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2022-11-16 Classification: DNA BINDING PROTEIN Ligands: K |
 |
Organism: Danio rerio
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2022-11-16 Classification: DNA BINDING PROTEIN Ligands: ACT |
 |
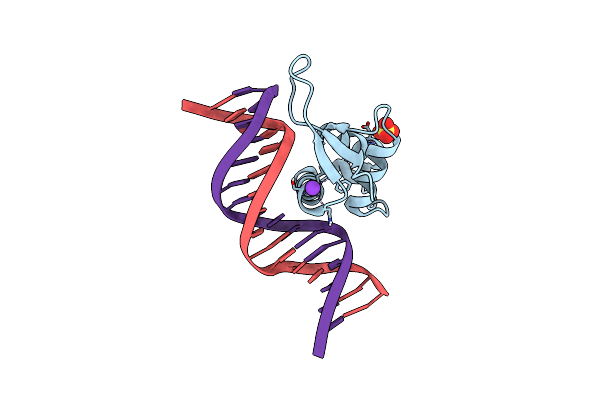
Crystal Structure Of The Zebrafish Foxh1 Bound To The Tgtttact Site (Fkh Motif Gtaaaca)
Organism: Danio rerio
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2022-11-16 Classification: DNA BINDING PROTEIN Ligands: ACT, PGE |
 |
Crystal Structure Of The Human Foxa2 Bound To The Tgtttact Site (Forkhead Motif Gtaaaca)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2022-11-16 Classification: DNA BINDING PROTEIN Ligands: K, SO4 |
 |
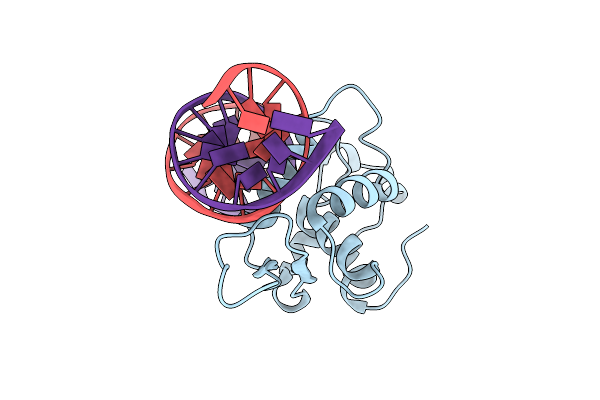
Crystal Structure Of The Human Foxa2 Bound To The Tgtttatt Site (Forkhead Motif Ataaaca)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2022-11-16 Classification: DNA BINDING PROTEIN Ligands: K |
 |
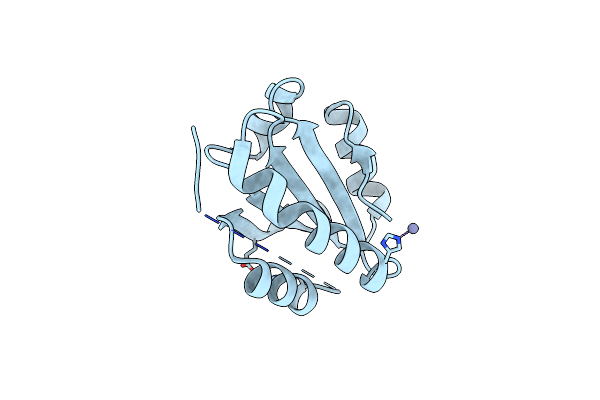
Organism: Xenopus laevis
Method: X-RAY DIFFRACTION Resolution:2.82 Å Release Date: 2022-11-16 Classification: DNA BINDING PROTEIN |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2020-10-21 Classification: NUCLEAR PROTEIN Ligands: ZN |
 |
Crystal Structure Of The Germline-Specific Thioredoxin Protein Deadhead (Thioredoxin-1) From Drospohila Melanogaster, P43212
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Release Date: 2020-10-21 Classification: OXIDOREDUCTASE Ligands: SO4, NA |
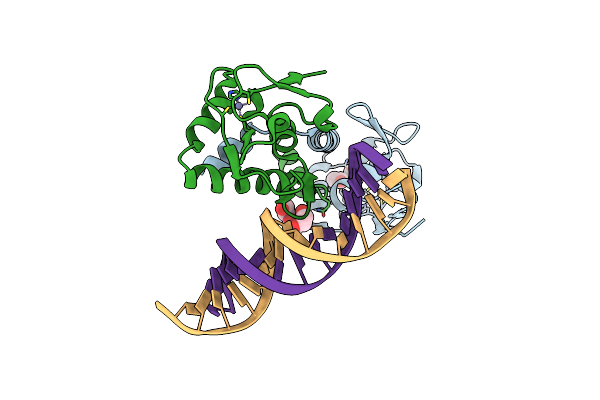 |
Crystal Structure Of The Smad3-Smad5 Mh1 Domain Chimera Bound To The Ggcgc Site
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2020-08-26 Classification: SIGNALING PROTEIN Ligands: ACT, EDO, ZN, PGE |
 |
Crystal Structure Of The Mh1 Domain Of Smad5-Smad3 Chimera Construct Bound To The Ggcgc Site
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2019-11-20 Classification: TRANSCRIPTION Ligands: ZN |
 |
Crystal Structure Of The Ggct Site-Bound Mh1 Domain Of Smad5 Containing A Gggs Insertion In The Loop1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.92 Å Release Date: 2019-11-20 Classification: TRANSCRIPTION Ligands: ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2019-10-02 Classification: TRANSCRIPTION Ligands: ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2019-10-02 Classification: TRANSCRIPTION Ligands: ZN, GOL |
 |
Organism: Streptococcus agalactiae, Plasmid pmv158
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-04-12 Classification: DNA BINDING PROTEIN Ligands: CL, MG, GOL, NA |
 |
Mobm Relaxase Domain (Mobv; Mob_Pre) Bound To Plasmid Pmv158 Orit Dna (22Nt). Mn-Bound Crystal Structure At Ph 4.6
Organism: Streptococcus agalactiae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2014-09-24 Classification: DNA BINDING PROTEIN/DNA Ligands: MN, GOL |
 |
Mobm Relaxase Domain (Mobv; Mob_Pre) Bound To Plasmid Pmv158 Orit Dna (22Nt). Mn-Bound Crystal Structure At Ph 5.5
Organism: Streptococcus agalactiae
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2014-09-24 Classification: DNA BINDING PROTEIN/DNA Ligands: MN, ACT |
 |
Mobm Relaxase Domain (Mobv; Mob_Pre) Bound To Plasmid Pmv158 Orit Dna (22Nt+3'Phosphate). Mn-Bound Crystal Structure At Ph 4.6
Organism: Streptococcus agalactiae
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2014-09-24 Classification: DNA BINDING PROTEIN/DNA Ligands: MN, NA |

