Search Count: 41
 |
Organism: Drosophila melanogaster
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: VIRUS LIKE PARTICLE |
 |
Cryo-Em Structure Of Single-Layered Nucleoprotein-Rna Helical Assembly From Marburg Virus, Trimeric Repeat Unit
Organism: Marburg virus - musoke, kenya, 1980
Method: ELECTRON MICROSCOPY Release Date: 2024-10-09 Classification: VIRAL PROTEIN |
 |
Structural Basis For Vipp1 Oligomerization And Maintenance Of Thylakoid Membrane Integrity
Organism: Synechocystis sp. (strain pcc 6803 / kazusa)
Method: ELECTRON MICROSCOPY Release Date: 2021-06-30 Classification: LIPID BINDING PROTEIN Ligands: ADP |
 |
Structural Basis For Vipp1 Oligomerization And Maintenance Of Thylakoid Membrane Integrity
Organism: Synechocystis sp. (strain pcc 6803 / kazusa)
Method: ELECTRON MICROSCOPY Release Date: 2021-06-30 Classification: LIPID BINDING PROTEIN Ligands: ADP |
 |
Structural Basis For Vipp1 Oligomerization And Maintenance Of Thylakoid Membrane Integrity
Organism: Synechocystis sp. (strain pcc 6803 / kazusa)
Method: ELECTRON MICROSCOPY Release Date: 2021-06-30 Classification: LIPID BINDING PROTEIN Ligands: ADP |
 |
Structural Basis For Vipp1 Oligomerization And Maintenance Of Thylakoid Membrane Integrity
Organism: Synechocystis sp. (strain pcc 6803 / kazusa)
Method: ELECTRON MICROSCOPY Release Date: 2021-06-30 Classification: LIPID BINDING PROTEIN Ligands: ADP |
 |
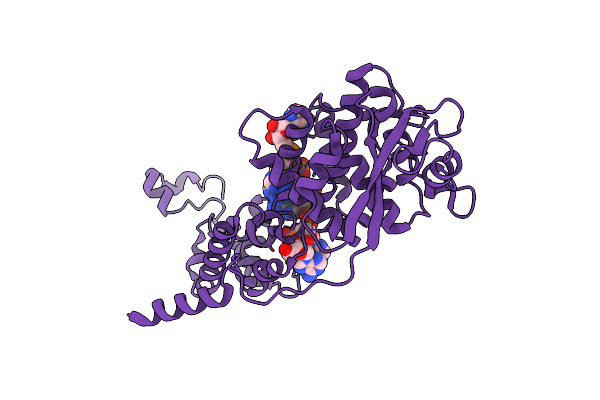
Structural Basis For Vipp1 Oligomerization And Maintenance Of Thylakoid Membrane Integrity
Organism: Synechocystis sp. (strain pcc 6803 / kazusa)
Method: ELECTRON MICROSCOPY Release Date: 2021-06-30 Classification: LIPID BINDING PROTEIN Ligands: ADP |
 |
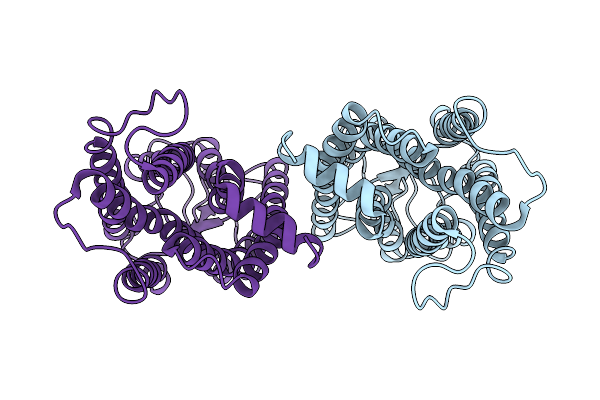
Organism: Morbillivirus sp.
Method: ELECTRON MICROSCOPY Release Date: 2021-06-23 Classification: VIRAL PROTEIN |
 |
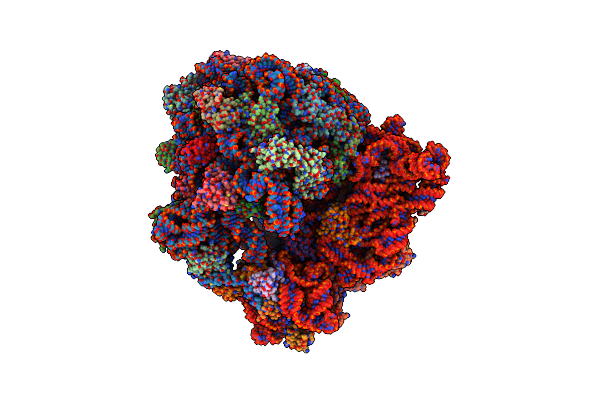
Cryo-Em Structure Of The Native Rhodopsin Dimer From Rod Photoreceptor Cells
Organism: Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2019-08-21 Classification: SIGNALING PROTEIN |
 |
Organism: Lama glama, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2018-09-19 Classification: SIGNALING PROTEIN |
 |
Structure Of A Hibernating 100S Ribosome Reveals An Inactive Conformation Of The Ribosomal Protein S1 - 70S Hibernating E. Coli Ribosome
Organism: Escherichia coli bw25113
Method: ELECTRON MICROSCOPY Release Date: 2018-09-05 Classification: RIBOSOME |
 |
Structure Of A Hibernating 100S Ribosome Reveals An Inactive Conformation Of The Ribosomal Protein S1 - Full 100S Hibernating E. Coli Ribosome
Organism: Escherichia coli bw25113
Method: ELECTRON MICROSCOPY Release Date: 2018-09-05 Classification: RIBOSOME |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2018-08-29 Classification: HYDROLASE Ligands: ATP, MG, ADP |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2018-08-22 Classification: HYDROLASE Ligands: ATP, MG, ADP |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2018-08-22 Classification: HYDROLASE Ligands: ATP, MG, ADP |
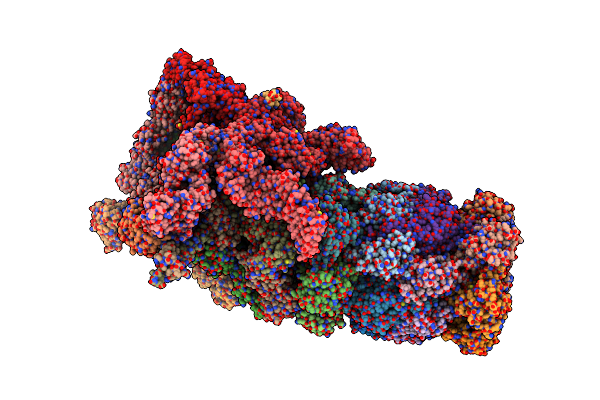 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2018-08-22 Classification: HYDROLASE Ligands: ADP, MG, ATP |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2018-08-22 Classification: HYDROLASE Ligands: ATP, MG, ADP |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2018-08-22 Classification: HYDROLASE Ligands: ATP, MG, ADP |
 |
Cryo-Em Structure Of The Human Adenosine A1 Receptor-Gi2-Protein Complex Bound To Its Endogenous Agonist
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2018-06-20 Classification: SIGNALING PROTEIN Ligands: ADN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2017-09-13 Classification: TRANSCRIPTION Ligands: GOL, CL |

